Research papers
← vista completaPublished on December 4, 2021 | http://doi.org/10.5867/medwave.2021.11.002099
Protective association exhibited by a single nucleotide polymorphism of the IFIH1 gene in patients with psoriasis: A case-control study
Asociación protectora del polimorfismo de nucleótido único en IFIH1 en pacientes con psoriasis: estudio de casos y controles
Abstract
Introduction Psoriasis is a chronic inflammatory dermatosis, a with variable clinical presentation and whose multifactorial etiology carries an essential genetic component. Multiple genetic variations associated with psoriasis have been described around the world. However, these variants are unknown among the Colombian population. This study aimed to evaluate the single nucleotide polymorphism rs10930046 (His460Arg) in the IFIH1 gene and its ssociation with the development of psoriasis in a Colombian population.
Methods An observational, unmatched, case-control study was performed, including 51 patients with psoriasis and 151 population controls, all with self-reported Paisa ancestry (from the Antioquia region). All individuals were genotyped for the single nucleotide polymorphism rs10930046 (His460Arg) in the IFIH1 gene, and its association with psoriasis was pursued. Both groups were demographically characterized, and cases were also assessed for clinical variables.
Results Through the allelic association analysis, cases were found to have a lower frequency of the single nucleotide polymorphism rs10930046 (His460Arg) in the IFIH1 gene than controls; 5% versus 22.67%, respectively. There were no significant differences in age or sex. We also found that psoriasis vulgaris was the most common variant (78%), that about half of the cases had nail psoriasis (56%), 19.6% had psoriatic arthritis, and that 45% had some comorbidity.
Conclusions The results obtained from this study confirm that carriers of the single nucleotide polymorphism rs10930046 (His460Arg) in the IFIH1 gene have a decreased risk of developing psoriasis.
Introduction
Psoriasis is a chronic inflammatory dermatosis whose worldwide incidence and prevalence range from 60.4 to 140 cases per 100 000 population and from 0.09 to 5.6%, respectively [1].
Different clinical subtypes include psoriasis vulgaris, guttate psoriasis, erythrodermic psoriasis, palmoplantar psoriasis, inverse psoriasis, and pustular psoriasis. These subtypes may or may not present ungular and/or joint involvement [2],[3]. The most common form of these clinical phenotypes is psoriasis vulgaris, accounting for up to 90% of all cases in some series [3]. Although the etiopathogenesis of psoriasis is not yet fully understood, it is known that – in individuals with some genetic predisposition – the disease is triggered by biochemical, immunologic, and vascular system abnormalities [2]. Additionally, psoriasis has been associated with several comorbidities such as inflammatory bowel diseases, cardiovascular syndrome, metabolic syndrome, depression, and cancer [4],[5].
As in any other autoimmune disease, the manifestation of psoriasis requires the combination of three components: genetic predisposition, environmental factors, and altered immune regulation.
The contribution of genetic determinants to psoriasis has been elucidated from epidemiological studies, genetic linkage studies [6],[7], and more recently from genome- wide association studies (GWAS) [6]. The latter has allowed the identification of single nucleotide polymorphisms (SNP, Simple Nucleotide Polymorphism) associated with the development of the disease [6],[8]. Most of them are located in genes involved in antigenic presentation processes (HLA- C and ERAP1), antiviral innate immune response (IFIH1, DDX58, TYK2, RNF114), and fundamentally, involved in Th17 immune response activation pathways (IL12B, IL23A, IL23R, TRAF3IP2, and NFKBIZ) [9],[10].
The IFIH1 (Interferon Induced with Helicase c domain 1), located on chromosome 2q24.3, is described among the associated genes. This gene encodes for the MDA5 protein and consist of three domains:
- An N-t erminal caspase recruitment domain (CARD, Caspase Recruitment Domain), initiating signaling cascades leading to the final activation of interferon regulatory factor (IRF) 3, IRF-7 , and NF-kB.
- A central RNA helicase domain with antiviral activity, triggering the activation of an adaptor protein, IFN-B promoter stimulator-1 (IPS-1) , and the production of IFN-B , among other pro- inflammatory cytokines.
- And a C- terminal domain [11],[12]. Single nucleotide polymorphisms in this gene could alter the function and/or decrease the concentration of the MDA5 protein in circulating mononuclear cells, reducing the production of pro-i nflammatory factors involved in the pathogenesis of autoimmune diseases [13], and conferring to its carrier a decreased risk of developing them [14],[15].
In a study performed in our population, variants in the IFIH1 gene, including the single nucleotide polymorphism rs10930046, were evaluated in a cohort of family trios with type 1 diabetes mellitus (T1D). The C allele of this single nucleotide polymorphism revealed a protective effect for developing T1D1 (205 cases versus 96 controls) [16]. Interestingly, in study of a Caucasian population, this allele showed the same effect in psoriasis (2098 cases versus 1744 controls) [10].
To date, no reports of genetic studies related to psoriasis have been found in Colombia. Thus, finding the single nucleotide polymorphism rs10930046 (IFIH1 gene) as a protective allele for type 1 diabetes in the Colombian population [16] encouraged us to elucidate its effect on psoriasis based on the fact that autoimmunity is implicated in both diseases. Therefore, this study aimed to evaluate the association of the single nucleotide polymorphism rs10930046 (His460Arg) in the IFIH1 gene with the development of psoriasis in Colombian patients.
Methods
Population
The design consisted of an unmatched case-c ontrol study, where the ratio of cases to controls was 1:3. Cases corresponded to patients with psoriasis and its information was collected prospectively (incident cases) during dermatologic care in different centers in Medellin, Colombia. The inclusion criteria of cases consisted of:
- Any clinical variant of psoriasis, diagnosed clinically and/or histopathologically by a dermatologist.
- Self-r eported paisa ancestry (well-characterized and genetical- ly studied Colombian population, predominantly located in Antioquia).
- Age equal to or older than 18 years.
Exclusion criteria for the cases corresponded to any mental deficit that prevented their participation in the study.
All patients signed an informed consent form. The study was framed within the norms for research in human subjects stipulated in resolution No. 8430 of 1993 of the Colombian Ministry of Health and the 2013 Declaration of Helsinki. The study was submitted to and approved by the ethics committee of the ‘Institución Prestadora de Servicios de Salud Universitaria, Hospital San Vicente Fundación’ and the ‘Instituto de Investigaciones Médicas’ from the School of Medicine, Universidad de Antioquia in Medellín, Colombia.
We verified paisa ancestry up to the second degree of consanguinity of cases, and a dermatologist evaluated and collected sociodemographic and clinical information of each patient. Early- onset psoriasis was defined as psoriasis presenting before 40 years of age. The Psoriasis Area and Severity Index (PASI) clinical scale was used to assess the severity of psoriasis. Nail psoriasis was defined by inherent disease changes in the ungular plaque on one or more nails. The presence of psoriatic arthritis was also assessed. Three groups were considered for family history of psoriasis: first, second, and third or higher degree of consanguinity.
Controls corresponded to individuals of self-r eported paisa ancestry, parents of children diagnosed with idiopathic generalized epilepsy, with no history of autoimmune diseases, and who had participated in a previous local genetic study. Demographic and genetic information (specifically the presence or absence of the single nucleotide polymorphism rs10930046 – p. His460Arg – in the IFIH1 gene) was obtained from that registry. Among controls, we excluded patients with a history of autoimmune disease and patients in whom a diagnosis of psoriasis was identified in the genetic registry.
Selection of the single nucleotide polymorphism
The single nucleotide polymorphism rs10930046 (His460Arg) was selected in the IFIH1 gene located on chromosome 2q24.3, where a missense substitution in the protein leads to the replacement of histidine by arginine at position 460 (available at http:/www. genome.u csc.edu). This substitution was chosen based on a study published in North America relating it to psoriasis as a protective factor in Caucasian Americans [10], and because this same polymorphism has been associated with the development of other inflammatory and autoimmune diseases [12],[16].
Genotyping
DNA was extracted from buccal cells using the modified salting-o ut method [17]. Genotyping was performed by the tetra-p rimer ARMS- PCR (Amplification Refractory Mutation System-P olymerase Chain Reaction) method on an Eppendorf Nexus X2 thermal cycler. The primers used were: 5'-T TTT AGCT TTCTT TGC TTTCA TTGGTTATACA- 3' (outer for- ward), 5'-T GTTTTTTT CAA CTTC TGCA TCAA ATCAT3' (inner forward), 5'-A AAG CTAT TTTCT CAC TCAC TGTA GTGTG TGCTG-3 ' (outer reverse) and 5'-G AAG CAGT GTGT ATAA TAAT AACA TCAT CATG AGTC GAGTCG- 3' (inner reverse). PCR conditions were: 94 degrees Celsius for five minutes; 35 cycles of 94 degrees Celsius for 45 seconds; alignment was performed at 56 degrees Celsius for 45 seconds followed by extension at 72 degrees Celsius for 45 seconds; and the final extension step at 72 degrees Celsius for 10 minutes. PCR products were directly analyzed by 3% agarose gel electrophoresis stained with EZVision (AMRESCO, SOLON, OH, USA), and each allele was recognized according to size. (Figure 1)
Sample size
According to Rodriguez and collaborators [16], and based on a prevalence of 0.31 of the single nucleotide polymorphism 460Arg (rs10930046) in the IFIH1 gene, we calculated a minimum sample of 45 cases and 135 controls sample considering a power of 80%, a confidence level of 90%, and an expected odds ratio of 0.41 (OpenEpi© Version 3.03.17) [18].
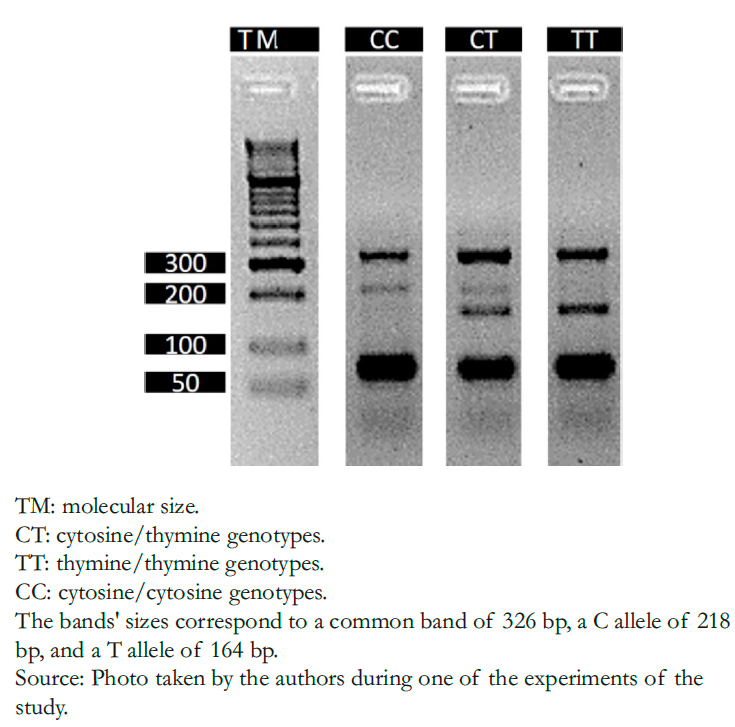 Full size
Full size Statistical analysis
Univariate analysis of the variables was performed to find their frequencies. The normal distribution of quantitative variables was evaluated with the Kolmogorov-S mirnov test. Multivariate logistic regression analysis was used to identify significant associations and the strength of association between exposure and response variables and verify potential confounders (e.g., presence of autoimmunity). In this regard, the standard measure of association in case-c ontrol studies (i.e., odds ratio or disparity ratio) was estimated with its respective 95% confidence interval. PLINK software version 1.9 was used to: calculate allelic frequencies, Hardy- Weinberg equilibrium (HWE), and the allelic association of rs10930046 with psoriasis. A P < 0.05 was considered statistically significant.
Results
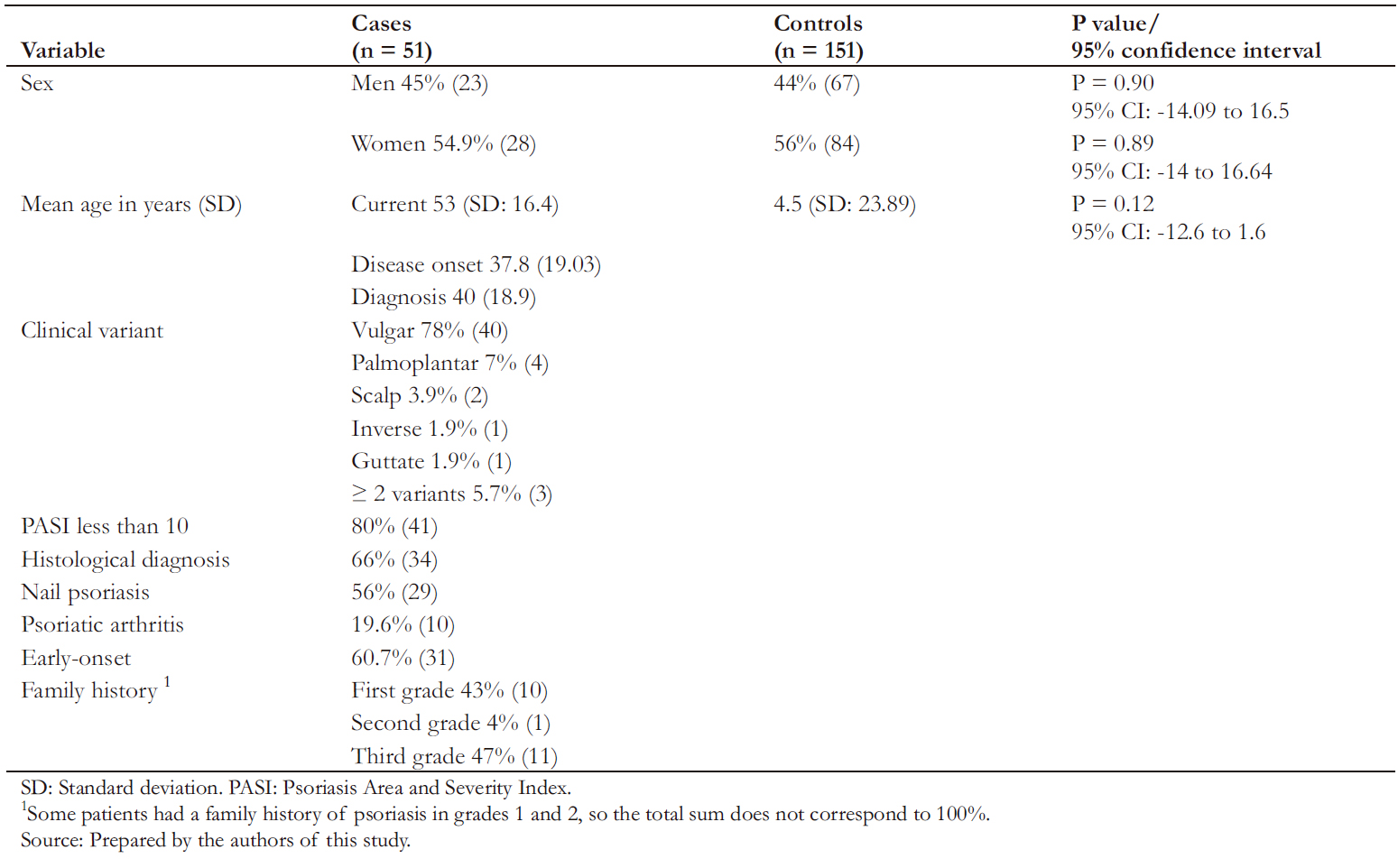
A total of 202 individuals were included, of which 51 were cases, and 151 were controls. Table 1 describes the sociodemographic characteristics of each group.
Demographic characterization
All individuals in the study had self-r eported Paisa ancestry. The "paisa" population corresponds to a particular ethnic distribution in Colombia with a high consanguinity prevalence and geographic isolation.
The mean age of cases and controls was 53 years (standard deviation ± 16.4) and 47.5 years (standard deviation ± 14.7), respectively. Among cases, men represented 45% of the sample. No statistically significant differences were found in sex or age among groups (Table 1).
Clinical characterization of cases
Among cases, age at disease onset and diagnosis was 37.8 (standard deviation ± 19) and 40 (standard deviation ± 18.9) years, respectively. Early-o nset psoriasis corresponded to 60.7% of cases (48% males and 51% females). The most common clinical variant was psoriasis vulgaris (Table 2). As for the classification of psoriasis severity, the mild to moderate forms of the disease were the most frequent (80%, 41/51).
Nail psoriasis was found in 56% (27/51) of cases, and the presence of ungular changes without psoriatic arthritis was more common than with psoriatic arthritis. However, of the 19.6% (10/51) patients who were found to have psoriatic arthritis, the majority (60%, 6/10) had concomitant ungular changes attributable to psoriasis.
Table 3 shows that most frequent comorbidities found in the studied population corresponded to arterial hypertension (HTN) and type 2 diabetes mellitus.
 Full size
Full size 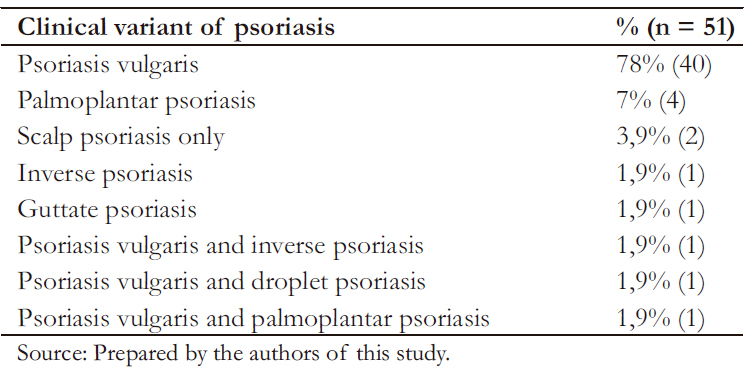 Full size
Full size 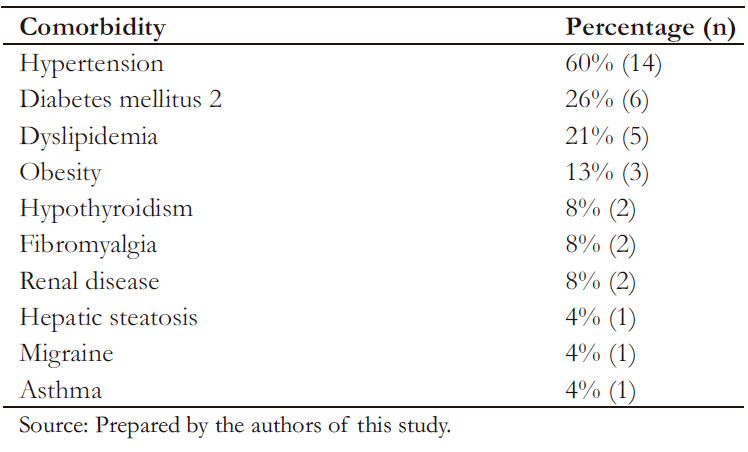 Full size
Full size Genetic analysis
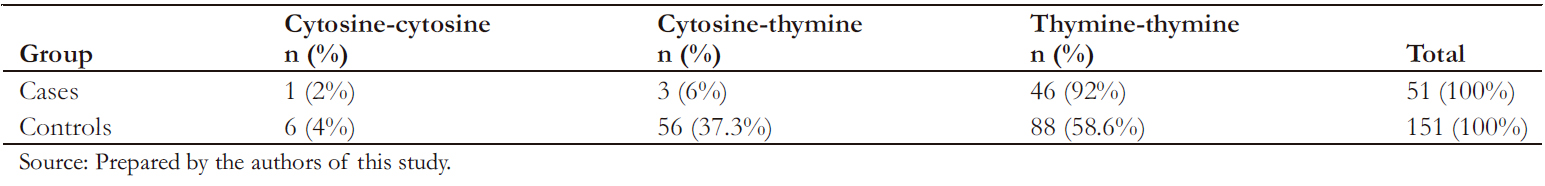
The genotyping rate was 99% for cases (50/51) and controls (150/151). Table 4 shows the genotypic frequencies found for the rs10930046 marker in the IFIH1 gene.
As shown in Table 4, the marker was in Hardy-W einberg equilibrium in the control group. The allelic association analysis of the C allele of rs10930046 with psoriasis was statistically significant (odds ratio: 0.17; 95% confidence interval: 0.07 to 0.45; P < 0.05), demonstrating that this allele confers significant protection against the development of psoriasis to its carrier.
A second allelic association analysis was performed to confirm results, comparing cases with genotypes of 96 healthy individuals, older than 18 years, of paisa ancestry, publicly available in the 1000 genomes database [19]. Through this analysis, a significant association of the C allele with psoriasis was observed (odds ratio: 0.17, 95% confidence interval: 0.06 to 0.46, P < 0.05) (Table 5).
In addition, to compensate for the lack of information from controls regarding personal history of psoriasis, an additional association analysis was performed based on the highest prevalence figure reported in the world (5.6%), which continued to show statistical significance.
 Full size
Full size 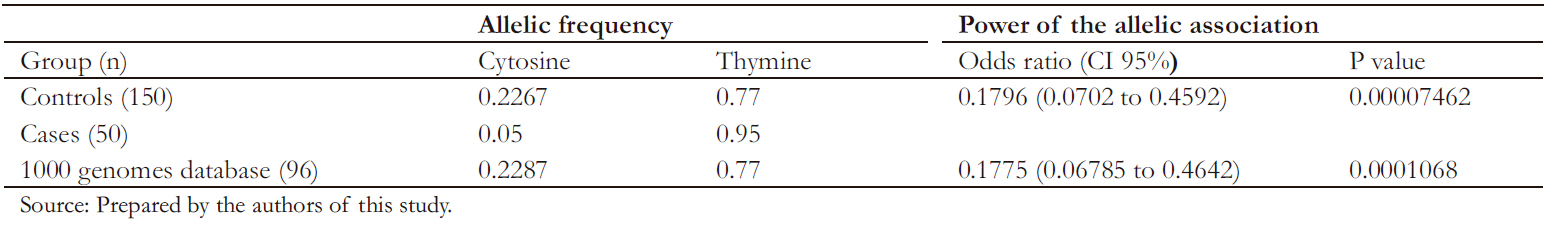 Full size
Full size Discussion
The results of this study suggest that the single nucleotide polymorphism rs10930046 in the IFIH1 gene modifies the risk of developing psoriasis in the Colombian population with paisa ancestry. Moreover, we found similarities in clinical and demographic characteristics of our sample with psoriasis patients in Colombia [20] and around the world [21],[22].
Similar to our findings, psoriasis has been found to predominantly affect patients over 18 years of age, being more common among 30 to 39 years and 50 to 69 years of age [21],[23]. Psoriasis has an early onset in 65.2% of cases [24], and psoriasis vulgaris is the most common variant. Likewise, we found similarities of clinical and demographic characteristics in our sample compared with other reports in patients with psoriasis in Colombia [20] and around the world [1],[3],[21],[22].
Regarding the existent body of evidence, there are no reports of population-b ased studies of psoriasis prevalence in Colombia or Latin America in general. A study found a global prevalence of psoriatic arthritis of 19.7% and in Latin America of 21.5% [1]. We found a prevalence similar to those numbers, where 19.6% had this condition. Furthermore, nail involvement has been reported in about 50% of patients with psoriasis [25], which resembles the 56% found in this study.
In Colombia, studies conducted in Bogotá reported mean ages at diagnosis of 46.3 and 54.5 years, with similar distribution between men and women [20]. These local epidemiological data are comparable with those reported worldwide, except for psoriatic arthritis, where a lower frequency is reported. This latter difference could be explained by underdiagnosis. However, we found that patients had prevalences of psoriatic arthritis similar to those reported in other parts of the world and other Latin American studies [26].
From a genetic point of view, multiple studies have identified genetic variants in patients with psoriasis, most of which code for molecules involved in immunological processes closely related to the disease [27]. In the IFIH1 (MDA5) gene, the following single nucleotide polymorphisms related to psoriasis have been described: rs17716942 [28], rs1990760 and rs35667974 [29], rs17715343 and rs13431841 [30].
Regarding the IFIH1 gene, only one study found an association between the single nucleotide polymorphism rs10930046 and the development of psoriasis [10]. This finding aligns with our results and confirms that this single nucleotide polymorphism confers a protective role for psoriasis to its carriers in these two specific populations, which could have some genetic similarities. Additionally, the protective role of this single nucleotide polymorphism for type 1 diabetes mellitus supports the hypothesis of a shared etiology for different forms of autoimmunity and inflammation processes [16].
One of the limitations found in the study was the lack of information regarding the presence or absence of psoriasis among controls, which corresponded to parents of children with idiopathic epilepsy. Another limitation was an insufficient sample size to find associations with clinical variants of the disease and comorbidities. Nevertheless, the power of the sample was sufficient to establish the protective genetic association of the single nucleotide polymorphism rs10930046 in the IFIH1 gene in patients with psoriasis in our population, which corresponded to our working hypothesis.
Among the study’s strengths, the rigorousness with which both the genetic tests were performed and the high methodological quality of the study stand out, which allowed us to minimize biases related to case-c ontrol studies.
Conclusion
We present the first genetic study on psoriasis carried out in the Colombian paisa population that confirms through allelic association analysis of cases and controls that carriers of the single nucleotide polymorphism rs10930046 (p. His460Arg) in the IFIH1 gene have a lower risk of developing psoriasis.
These results promote the development of a study with larger sample size. In this way, genetic findings could be compared with demographic and clinical variables, considering the low frequency of the C allele in the study population, as demonstrated in other genetic studies focused on the major histocompatibility complex [9],[31].
In all, genetic association studies such as this one allow determining characteristics related to the endotype of psoriasis. This approach may facilitate the identification and future development of effective therapies for patients with specific genetic characteristics. Additionally, it would benefit patients and health care systems, as it would ensure that each patient receives the appropriate treatment for their psoriasis.
Notes
Contributor roles
EVEL, CJES, DMCS: conceptualization, laboratory work, patient evaluation, results analysis, article writing, review, and editing. GS: conceptualization, methodology, patient evaluation, results analysis, software, visualization, supervision, project management, obtaining funding, article writing, review, and editing. NGP: conceptualization, methodology, laboratory work, results analysis, software, visualization, supervision, project management, obtaining funding, writing, reviewing, and editing article.
Acknowledgments
To Blear Holguín, laboratory assistant of the Genetic Mapping Research Group of the University of Antioquia, and Diana Cornejo, Ph.D. student of the Corporation of Basic Biomedical Sciences of the University of Antioquia. Special thanks to Mateo Cuervo, Daniela Gómez, Manuel Martínez, Natalia Vélez (Dermatology residents of the University of Antioquia), and the Phototherapy and Photodermatology Service of the Health Services Provider Institution of the University of Antioquia. Finally, we thank Alejandra Venegas, research assistant of the Grupo de Investigación Dermatológica (GRID) of the University of Antioquia, for contacting patients and managing informed consent.
Competing interests
The authors completed the ICMJE conflict of interest statement and declared that they received no funding for the completion of this article; have no financial relationships with organizations that may have an interest in the published article in the past three years; and have no other relationships or activities that may influence the publication of the article. Forms can be requested by contacting the responsible author or the Editorial Committee of the Journal.
Funding
The study was funded with resources from the Genetic Mapping Group and Grupo de Investigación Dermatológica (GRID), both attached to the Medical Research Institute of the School of Medicine of the University of Antioquia in Medellín, Colombia.
Ethics
This study was approved by the ethics committee of the ‘Institución Prestadora de Servicios de Salud Universitaria, Hospital San Vicente Fundación’ and the Research Institute of the School of Medicine of the University of Antioquia in Medellín, Colombia.
Data sharing statement
The database for this study is available upon request.
Provenance
Not commissioned.
Language of submission
Spanish.
Type of review
Externally peer-r eviewed by three reviewers, double-blind

