Clinical reviews
← vista completaPublished on September 26, 2020 | http://doi.org/10.5867/medwave.2020.08.8037
Origin, structural characteristics, prevention measures, diagnosis and potential drugs to prevent and COVID-19
Origen, características estructurales, medidas de prevención, diagnóstico y fármacos potenciales para prevenir y controlar COVID-19
Abstract
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) causes COVID-19 (Coronavirus disease 2019). This disease was detected in the city of Wuhan in China in December 2019. People infected with COVID-19 shows varying manifestations, depending on their health and age. The most common symptoms are fever, cough, myalgia, fatigue, odynophagia, and dyspnea. Infected adults older 60 years of age are the group of patients most susceptible to severe COVID-19 states and present comor-bidity in the presence of chronic diseases. On the other hand, it is also essential to have tests to detect SARS-COV-2 in people and follow the evolution of COVID-19 quickly, reliably, and cheap. To achieve this, there are real-time reverse transcription pol-ymerase chain reaction (RT-PCR) tests, isothermal nucleic acid amplification, and enzyme immunostimulation. Currently, there are no drug treatments to prevent infection and to combat the virus's effects. However, different research groups that are conduct-ing in vitro, in vivo, and in silico tests to find drugs able to provide an immune response and to control infection in humans with SARS-CoV-2. Chloroquine, hydroxychloroquine, remdesivir, interferon-2b, and oseltamivir are some pharmacological options evaluated in clinical trials for prophylaxis of COVID-19. The purpose of this review is to establish a reference framework for taxonomic classification of SARS-CoV-2 and the relationship they have with other CoVs, as well as their structure and propaga-tion pathways in humans. The characteristics and symptoms presented by patients with COVID-19, the detection methods, and possible treatments are also presented.
|
Main messages
|
Introduction
Currently, technological advances have generated connectivity never seen before, which has brought improvements in our living conditions and the appearance of new diseases and their spread, as is the case of Coronavirus Disease (COVID- 19) caused by SARS-CoV-2[1]. This virus is similar to SARS-CoV and MERS-CoV (MERS, the acronym for Middle East Respiratory Syndrome) CoVs that caused epidemics in this century[2],[3].
SARS-CoV-2 has spread since December 2019 from Wuhan (Hubei Province), China, to all of the most important cities worldwide. This has generated an unprecedented global health problem with a new zoonosis, which has marked the appearance of several information sources that are updated rapidly.
Symptomatic and asymptomatic persons of COVID-19 transmit SARS-CoV-2[4], and young patients with a median age of 22 years (from 16 to 23 years old) are the main asymptomatic transmitters[5]. Transmission occurs through droplets generated by coughs and sneezes. Fomites and other routes, such as fecal or air, are not ruled out[6]. Consequently, SARS-CoV-2 spreads quickly.
This article consists of a reference framework related to taxonomic classification, structure, and origin of SARS-CoV-2, as well as the factors that promoted its conversion into a pandemic. Information related to clinical diagnosis and tests being used for the detection of SARS-CoV-2 is also included.
Finally, some studies related to drugs with potential use for prevention and treatment of COVID-19 are presented with in vitro, in vivo, and in silicoapproaches. To fulfill this objective, we conducted a review of journals that regularly publish articles on coronaviruses, especially about SARS-CoV-2. The electronic pages of international organizations such as the World Health Organization (WHO), the Federal Drug Administration (FDA), and others were reviewed to a lesser degree.
Coronaviruses classification
Coronaviruses belong to the Nidovirales order and Coronaviridae family. They are classified according to their predominant genetic characteristics located within ORF1ab (pp1ab) replicase polyprotein. The most distinctive coronavirus characteristics are:
- The genome size is around 30 000 base pairs; coronaviruses are ribonucleic acid viruses with the largest genomes. This expansive coding ability appears to provide and necessitate many gene expression strategies.
- Expression of many non-structural genes by displacement of the ribosomal framework.
- Various unique or unusual enzyme activities are encoded within the large polyprotein replicase-transcriptase.
- Expression of downstream genes by synthesis of 3' nested subgenomic messenger ribonucleic acid, confirming close resemblance to host messenger ribonucleic acid.
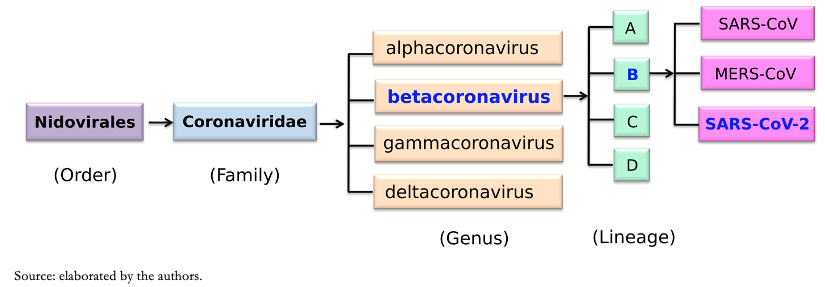
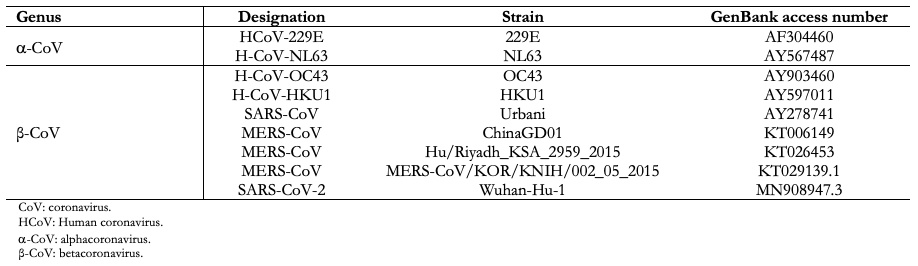
Nidovirales order name is derived from these nested 3' messenger ribonucleic acid. The main differences within nidovirus families are in the number, type, and size of structural proteins. Because viruses mutate quite easily, it has been established that strains isolated from different sources must have more than 90 percent homology (at the level of amino acids), within conserved replicase domains for them to be considered the same species[7]. Based on these criteria, coronaviruses, according to the Coronavirus Study group of International Committee on Taxonomy of Viruses, are classified into four genera, divided by phylogenetic groupings, alphacoronavirus (a-coronavirus), betacoronavirus (β-coronavirus), gammacoronavirus (γ-coronavirus) and deltacoronavirus (δ-coronavirus)[8]. Within genus β-coronavirus there are four lineages: lineages A, B, C, and D (Figure 1).
 Full size
Full size There are three main reasons for the great coronavirus diversity. First, coronavirus genomes are highly plastic because of the low diversity of their ribonucleic acid-dependent ribonucleic acid polymerases leads to high mutation rates of one per 1 000 to 10 000 replicated nucleotides[9],[10]. Second, coronaviruses have high rates of homologous ribonucleic acid recombination due to their unique mechanism of random template change during ribonucleic acid replication[11],[12]. Third, coronaviruses have a greater capacity to incorporate new genes than other ribonucleic acid viruses due to their relatively large genomes (from approximately 26 to 32 kilobases). These three factors allow diverse strains generation and genotypes within a single coronavirus species. Also, new variants or species can cause zoonotic outbreaks.
Coronavirus virions structure and SARS-CoV-2 action mechanism
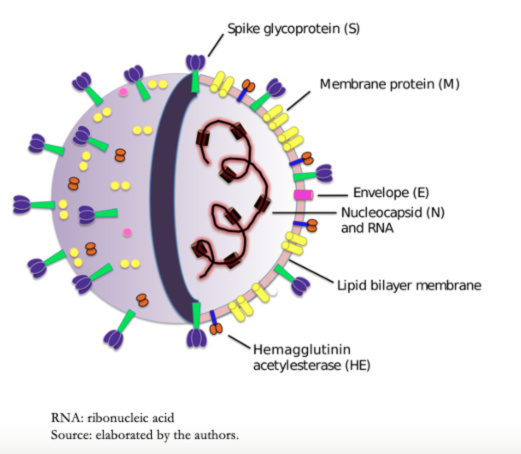
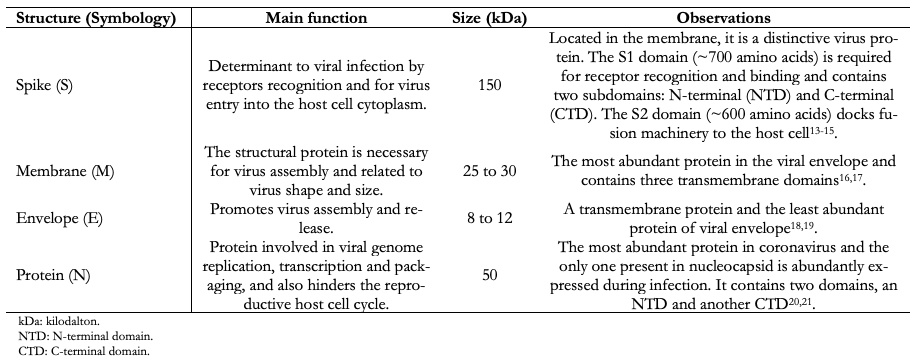
Coronavirus virions contain four major structural proteins. These are spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins. All these are encoded within the viral genome 3'-end (Figure 2 and Table 1). Coronaviruses are approximately spherical and moderately pleomorphic. Virions have generally been reported to have average diameters of 80 to 120 nanometers. Protein S (~ 150 kilodalton) uses an N-terminal signal sequence to gain access to the host. This protein forms a distinctive spike structure on the virus surface[13],[14]. S spikes project about 17 to 20 nanometers from the virion surface, with a thin base that swells to a width of about 10 nanometers at the distal end[15].
Protein M is a structural membrane glycoprotein located in the virion envelope. It is small (from 25 to 30 kilodalton) with three transmembrane domains[16] and plays a central role in virus assembly, turning cell membranes into factories where virus and host factors bind to produce new virions[17]. Protein E (from 8 to 12 kilodalton) is found in small amounts inside the virion. Proteins E are highly variable but have a common architecture[18]. Protein E has several functions, including to promote virus assembly and release, and this protein is necessary to infect new cells[19]. The nucleocapsid is only constituted by Protein N. This protein is constituted by two separate domains, an N-terminal domain, and a C-terminal domain, capable of binding to ribonucleic acid in vitro. However, each domain uses different mechanisms to bind to ribonucleic acid. Optimal ribonucleic acid-binding requires contributions from both domains[20],[21]. The β-coronaviruses show a second type of surface projection, five to ten nanometers long below the S spikes constituted by a homodimeric hemagglutinin-esterase glycoprotein, Figure 2. This protein is also a mediator of receptor binding. Viruses that express the hemagglutinin-esterase protein bind to erythrocytes and agglutinate this, and it is only found in β-coronavirus of lineage A[22],[23].
 Full size
Full size In most coronavirus, the protein S is cleaved by a host cell furin-like protease into two separate polypeptides denoted S1 and S2[23]. S1 constitutes the receptor-binding domain of protein S, allowing the virus to penetrate the host cell, while S2 contains the fusion mechanism, Figure 3.
 Full size
Full size 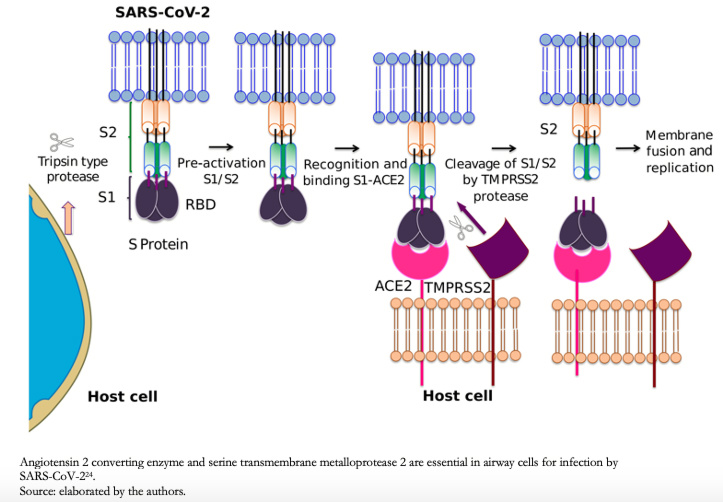 Full size
Full size Letko and Munster (2020) developed an approach to detect β-coronavirus of lineage B, such as SARS-CoV and SARS-CoV-2 rapidly, during receptor invasion and their ability to infect cells of different species types[25]. This study showed that host protease processing during viral entry is an important barrier for several β-coronavirus of lineage B, and by circumventing this barrier, viruses enter human cells through an unknown receptor. The authors also demonstrated how different β-coronavirus of lineage B could recombine to enter human cells and confirmed that human angiotensin-converting enzyme 2 is a SARS-CoV-2 receptor.
Human angiotensin-converting enzyme 2 is bound to the outer lung surface, arterial, heart, kidney, and intestinal cell membranes[26]. The SARS-CoV-2 S2 fragment, cleaved by a serine protease from the human cell surface, transmembrane metalloprotease serine 2, Figure 3, conduct to membrane fusion. According to Hoffmann and collaborators (2020), angiotensin-converting enzyme 2, and transmembrane metalloprotease serine 2 are essential in airway cells for SARS-CoV-2 infection[24].
Concerning angiotensin-converting enzyme 2, it is known to be overexpressed in patients with hypertension, type 1, and type 2 diabetes treated with angiotensin-converting enzyme inhibitors type-1 angiotensin II receptor blockers[27]. This condition can generate in patients with COVID-19, a more severe clinical course that involves damage to the myocardium, which has produced speculation since the association between angiotensin-converting enzyme 2 overexpression and infection risk or severity with SARS-CoV-2 is not fully understood[28].
The study reported by Chen and collaborators (2020) can help understand the relationship between patients with cardiovascular diseases and COVID-19. This study reported that of the 99 confirmed COVID-19 patients admitted to Wuhan Jinyintan Hospital, thirteen percent had elevated creatine kinase, and 76 percent demonstrated an elevated lactate dehydrogenase concentration[29]. Creatine kinase is found mainly in skeletal muscles and myocardium, and to a lesser extent, in the brain. A high concentration of creatine kinase may indicate damage to one of the muscles mentioned above. Like creatine kinase, lactate dehydrogenase is used to identify tissue lesions, including myocardium; however, lactate dehydrogenase is less specific than creatine kinase.
A very sensitive and specific myocardial injury marker is troponin I, which was evaluated by Wang and collaborators (2020) in 138 hospitalized COVID-19 patients at Wuhan University Zhongnan Hospital. Wang and collaborators (2020) found elevated troponin I levels in 7.2 percent of patients[30]. It should be noted that myocardial dysfunction can be indirectly caused by COVID-19, such as induced by reduced oxygen supply, severe lung failure, and a cytokine storm after SARS-CoV-2 infection. However, myocardial dysfunction is also a possible attribute to decreased angiotensin-converting enzyme 2 activity in the heart. At present, it is not yet feasible to distinguish all possible myocardial dysfunction promoters.
Origin of the coronaviruses that infect humans
In recent times, at least four zoonotic jump events of coronavirus from animals to humans have been recorded, resulting in epidemics. The first event occurred with a β-coronavirus from lineage A that involved at least nine animal species, including humans. It is speculated that the final zoonotic jump of this β-coronavirus was from bovines to humans about 120 years ago, leading to CoV OC43. This virus and H-CoV-229E were isolated and identified in 1967 and 1966, respectively. CoV OC43 and H-CoV-229E were the first coronaviruses identified in humans. For forty years, these coronaviruses were thought to be the only viruses that could infect humans[31]. Studies in healthy adult volunteers revealed that infection with these two coronaviruses can cause common cold symptoms. Additionally, for individuals infected with H-CoV-229E, rhinitis occurs, while for patients infected with H-CoV-OC43, odynophagia occurs. Therefore, these two viruses do not represent a serious health problem in the world.
The second zoonotic jumping event occurred at the beginning of the twenty-first century and involved a β-coronavirus of lineage B giving rise to human SARS-CoV in 2003. This event involved bats, palm civets (Paguma larvata), and humans, with the virus jumping from palm civets to humans[32]. Palm civet is a highly valued food in China. SARS-CoV was responsible for the epidemic that began in Guandong, China, in 2003[33]. This first pandemic of the twenty-first century created by coronavirus was controlled that same year by implementing quarantines.
The third zoonotic event occurred in β-coronavirus of lineage C, resulting in the middle east respiratory syndrome epidemic[34]. MERS-CoV is closely related to CoV HKU4 present in the Tylonycteris bat (Ty-BatCoV HKU4) and CoV HKU5 present in the Pipistrellus bat (Pi-BatCoV HKU5). Both species originated in Hong Kong[35]. On the other hand, outbreaks of HCoV-NL63 and H-CoV-HKU1 viruses appeared in 2004 and 2005, respectively. These latest human coronaviruses are not new; they just had not been detected previously. Additionally, there is evidence that H-CoV-HKU1 strain genotypes are recombined naturally[36].
The four human coronavirus strains mentioned above are classified as common cold viruses that, in some cases, severely affect young children, chronically ill adults, and the elderly. Another β-coronavirus of lineage C was responsible for the respiratory infection in humans that caused the Middle East respiratory syndrome in 2012 (MERS-CoV). At present, this β-coronavirus is well adapted to humans[37]. Therefore, at first glance, it appears that bats are the natural hosts of coronavirus that originated lineage C of β-coronavirus. However, a significant number of infections were not related to direct exposure to bats[38]. A more likely scenario is that a single variant of β-coronavirus in bats successfully jumped and established rapidly in an intermediate animal host species, dromedaries, and civets, with subsequent incidental spread to the human population. Such zoonotic events are facilitated through frequent human-intermediate host interactions and perhaps through viral adaptations acquired during the initial jump to the first species[35]. Therefore, animal sources and transmission routes between species during β-coronavirus of lineage C generation are still unknown.
The fourth zoonotic event corresponds to SARS-CoV-2, which is almost impossible to have originated from laboratory manipulation of a related coronavirus. This is because the receptor-binding domain in SARS-CoV-2 spike protein (S) is optimized to human ACE-2 bind differently from another coronavirus. Therefore, there are only two possible theories that can explain the SARS-CoV-2 origin:
- Natural selection in a host animal before zoonotic transfer to humans.
- Natural selection in humans after zoonotic transfer[39].
To date, no animal-hosted coronavirus has been identified that is similar enough to affirm that it is the direct precursor to SARS-CoV-2 unequivocally. However, mutations, which are insertions and deletions near S1-S2 protein complex junction, may occur in coronaviruses, and this may be evidence of a natural evolutionary process[40]. For a coronavirus hosted in an animal to acquire cleavage site from mutations in protein S to bind to human angiotensin-converting enzyme 2, the animal host must be in an environment with a high population density to promote efficient natural selection and for the coronavirus to express a gene that encodes angiotensin-converting enzyme 2 similar to human orthologous.
On the other hand, according to the second theory, it is also possible that a SARS-CoV-2 precursor has made a zoonotic jump to humans, acquiring genomic characteristics described above through adaptation during undetected transmission from human to human. Once these adaptations were acquired, the spread may intensify, and later a group of cases large enough to activate a surveillance system that detected it would be generated[41].
The reported SARS-CoV-2 genomes indicate that all isolates come from a common ancestor. Additionally, the presence of a receptor-binding domain like SARS-CoV-2 in pangolins suggests that the virus probably jumped from these animals to humans. SARS-CoV-2 has a ninety-one percent similarity to the Malayan pangolin β-Coronavirus at the total genome level and an aminoacids sequence similarity of 97.4 percent at the receptor-binding domain of protein S1[42]. SARS-CoV-2 and Malayan pangolin's (M. javanica) coronavirus have five identical amino acid residues that are key in coronavirus interaction with host receptors. Zhang and collaborators (2020) reached the same conclusions, noting that five key amino acid residues of S1 protein from SARS-CoV-2 involved in the interaction with human angiotensin-converting enzyme 2 are entirely consistent with those from Malayan pangolin's coronavirus[43]. These results allow having a high certainty that the Malayan pangolin is a SARS-CoV-2 reservoir.
In addition to all the above, coronaviruses evolve and present genetic recombinations either within the host species or when jumping from one species to another. These recombinations between different coronavirus strains can lead to a mutated coronavirus with a high degree of pathogenicity and can become potentially more fatal to humans[44]. The SARS-CoV genome's mutation rate was estimated at 0.80-2.38 × 10-3nucleotide substitutions per site per year[45]. In SARS-CoV-2, two mutations of protein S and protein N may explain its zoonotic transmission[46].
In general, comparative analysis of viral genomic sequences is useful to trace the coronavirus origin that infects humans and determines the relationship with other analogous viruses[47]. From these studies, it has been proven that almost all coronaviruses from a-coronavirus and β-coronavirus groups are hosted in mammals, and human coronaviruses are in both groups, Table 2. In contrast, γ-coronavirus have been isolated only from avian hosts. The δ-coronavirus has been isolated from various species of songbirds and leopard cats in China and pigs[48].
 Full size
Full size SARS-CoV-2 turned into a pandemic
Ribonucleic acid viruses, such as SARS-CoV-2, tend to favor pandemics due to the evolutionary plasticity inherent in ribonucleic acid genomes and disruption that humans induce in ecosystems where viruses occur naturally. Also, ribonucleic acid viruses have short generation times and fast evolutionary rates[49]. Rapid evolutionary rates of ribonucleic acid viruses develop from frequent replication cycles. This replication is susceptible to errors and can produce large populations of closely related genotypes[50]. Furthermore, without a commercial treatment available to contain the viral outbreak, and without a vaccine, it forces containment efforts at least during the early spread stages[51].
SARS-CoV-2 is responsible for the so-called COVID-19 disease. The emergence of COVID-19 is attributed to live animal seafood sales and wild animals in Wuhan, China. Prevailing conditions at the site possibly caused a zoonotic event.
The SARS-CoV-2 spread worldwide can be explained by the basic reproductive number (R0) of the virus, around 2.5[52]. This value indicates that one person will infect 2.5 people in the absence of preventive measures. For an outbreak to occur, the R0 value must be greater than unity. R0 for SARS-CoV-2 in China in epidemic early stages was 2.5, and based on this value, Anderson et al. (2020) calculated that in the absence of mitigation measures, approximately 60% of the world's population would be infected[52]. This is the most catastrophic scenario, however, in reality, there are populations less exposed than others, and in the countries most affected by the pandemic, contagion preventive measures have been implemented. These actions will reduce the SARS-CoV-2 spread significantly. Also, it is important to mention that as the pandemic progresses, the effective reproduction factor (Re) will decrease until it reaches values below unity, which will indicate pandemic control. Re's value is strongly dependent on control measures. Data reported by Kissler and collaborators (2020) suggest that Re has decreased to values of around 1.4[53]. Despite this being an encouraging result, it is important to note that social distancing measures may require several months to effectively control transmission and reduce the possibility of a SARS-CoV-2 outbreak. To understand the impact of such measures, Koo, Cook, and Park (2020) developed a model to project potential intervention effects on SARS-CoV-2 spread in Singapore[54]. These results show that starting with an R0 of 2.5 and considering a combined intervention based on quarantine, school closings, and physical distancing in the workplace, an effectiveness of 78.2 percent can be reached in reducing virus spread. In addition, China's results to contain the SARS-CoV-2 spread confirm that quarantining, social distancing, and isolation of infected populations can effectively contain the pandemic[55]. These measures can help reduce contagion in people who are highly susceptible to SARS-CoV-2. Chen and collaborators (2020) carried out a study where it becomes evident that older adults with chronic comorbidities, such as cerebrovascular, cardiovascular, endocrine, digestive, respiratory, nervous system diseases, and malignant tumors, may become more susceptible to being infected and have a severe state of COVID-19[29].
All ages are susceptible to becoming infected with SARS-CoV-2. This virus is transmitted through droplets generated by coughs and sneezes by symptomatic and asymptomatic patients[56]. Studies have shown higher viral loads in the nasal cavity than in the throat, with no viral load differences between symptomatic and asymptomatic people[57]. Patients can infect during the duration of symptoms and even in clinical recovery. Drops produced by coughing and sneezing can reach distances of one to two meters and settle on surfaces. SARS-CoV-2 can remain viable on surfaces during days in favorable atmospheric conditions, but it is quickly destroyed by common disinfectants such as sodium hypochlorite, hydrogen peroxide, among others[58]. The infection is acquired either by inhaling these droplets or by exposure to surfaces contaminated with the virus. SARS-CoV-2 is also present in feces, and it is speculated that contamination of the water supply may be a transmission source[59].
Even though most countries in the world have taken social distancing measures, data published on July 16, 2020, by the Coronavirus Resource Center at the University of Medicine Johns Hopkins on COVID-19 reported 13 758 533 people infected and 589 093 deaths worldwide, yielding a case fatality rate of 4.3 percent. Some European countries, such as Great Britain, have a case fatality rate of 15.4 percent, which is high compared to the global average, while other European countries have a relatively low rate, such as Germany, which has a case fatality rate of 4.5 percent. In the Americas, the United States has the highest death toll globally but with a case fatality rate of 3.9 percent. In Latin America, Brazil has the highest number of infected at 2 012 151 and deaths with 76 688, with a case fatality rate of 3.8 percent. Mexico has 324 041 infected and 37 574 deaths, with a case fatality rate of 11.6 percent. After Mexico, Ecuador has the highest case fatality, at 7.3 percent, in Latin America. Finally, Chile, the third country in Latin America with the highest number of infected, reports 323 698 cases; however, this country's case fatality is low compared to the global average at 2.2 percent. It is important to note that the case fatality rate makes it possible to assess disease behavior at a given point in the pandemic and varies with time. The case fatality rate reported here was calculated with the data available as of July 16, 2020.
COVID-19 characteristics and symptoms
CoVID-19 can occur in three possible states: mild, moderate, and severe. Patients under 60 years of age and without relevant comorbidities can present the disease in a mild or moderate state, while patients with significant comorbidities may present a moderate state. Moderate cases also present symptoms of pneumonia[60]. Adult patients with a severe COVID-19 state manifest tachypnea (³30 breaths•min-1), £ 93% oxygen saturation at rest, the PAFI index defined as the ratio of partial arterial pressure of oxygen (PaO2)/inspired oxygen fraction (FiO2)-1 £ 300 mm Hg (if the patient is found 1000 m above sea-level, the formula must be corrected)[60].
The WHO recommends collecting samples from both the upper respiratory tract (nasal and oropharyngeal samples) and the lower respiratory tract, expectorated sputum, endotracheal aspirate, or bronchoalveolar lavage. Bronchoalveolar lavage samples collection should only be performed in mechanically ventilated patients, as lower respiratory tract specimens appear to remain positive for a more extended period. Samples require storage at 4ºC. At this stage of identifying patients with COVID-19, it becomes important to use reliable laboratory tests for the disease diagnosis. These tests usually consist of reverse transcription polymerase chain reaction or isothermal nucleic acid amplification assays, which will be described later.
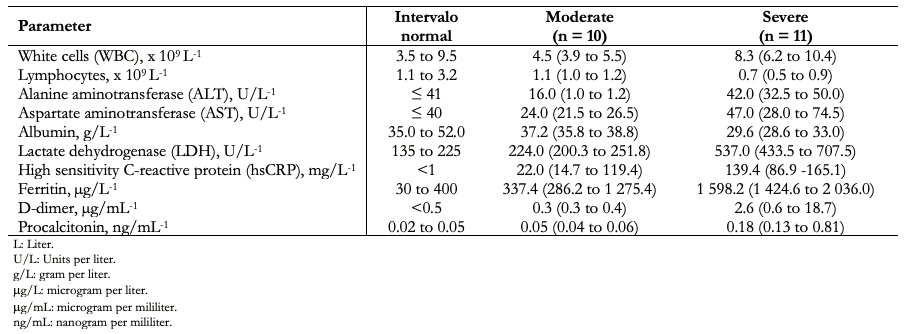
The clinical manifestations of patients with COVID-19 include fever, fatigue, dry cough, shortness of breath, and acute respiratory distress syndrome. White blood cell count is usually normal or low. There may be lymphopenia; a lymphocyte count < 1000 has been associated with a more severe condition; the platelet count is usually normal or slightly low. C-reactive protein and erythrocyte sedimentation rate are generally elevated, but procalcitonin levels are usually normal. A high level of procalcitonin can indicate a bacterial coinfection. Alanine aminotransferase, aspartate aminotransferase, prothrombin time, creatinine, D-dimer, creatine phosphokinase, and lactate dehydrogenase may be elevated, and high levels are associated with a severe disease state. Table 3 lists some clinical laboratory parameters of twenty-one patients studied in China classified as moderate (ten cases) and severe (eleven cases), according to the National Commission of China[61]. It should be noted that these symptoms and the clinical laboratory results are not entirely decisive to clarify the severity of COVID-19 patients.
 Full size
Full size In addition to blood parameters described in Table 3, patients suspected of COVID-19 should undergo a clinical review that includes taking vital signs, a routine chest examination, and oxygen saturation measurement using an oximeter. It should be noted that COVID-19 cannot be differentiated clinically or through routine laboratory tests from respiratory viral infections, such as influenza, parainfluenza, respiratory syncytial virus, among others. Additionally, patients with moderate or severe COVID-19 should ideally undergo a high-resolution chest computed tomography scan to assess the degree of involvement. Chest computed tomography radiological images combined with clinical manifestations and anatomopathological verification confirm the COVID-19 pulmonary pathology. The chest computed tomography scan evaluation of a patient with COVID-19 should consider the following characteristics[62]:
- “Ground glass” opacities presence.
- Consolidation presence.
- Laterality of opacities and consolidation in ground glass opacities.
- Number of affected lobes where ground glass opacities or consolidated were present.
- Involvement degree of each lung lobe in addition to overall degree of lung involvement as measured by a "total severity score"[63].
- nodules presence.
- Pleural effusion presence.
- Thoracic lymphadenopathy presence (from a lymph node size ≥10 mm in short axis dimension).
- Airway abnormalities.
- Disease axial distribution.
- Underlying lung disease presence, such as emphysema or fibrosis.
Typical findings of chest computed tomography images in patients with COVID-19 are bilateral multifocal opacities of irregular “ground glass” type or consolidation with septal and vascular interlobar thickening, mainly in lungs peripheral fields. Ground glass opacities is a cloudy area of increased lung attenuation with preserved bronchial and vascular markings. In patients with COVID-19, ground-glass opacities incidence rates and consolidation are approximately 86 percent and 29 percent, respectively[64]. The most ground glass opacities common in morphology are irregular and round, followed by triangular and linear[65]. Wu et al. (2020) mention that triangular ground glass opacities under the pleura with interlobular septal thickening, called "cobweb", are considered another indication of COVID-19 infection.
CT findings of COVID-19 patients may change as the disease progresses. In the early stage, chest computed tomography shows small irregular lobar and subsegmental ground-glass opacities, interstitial changes, and thickening of vascular lumens throughout. At a later and peak stage, lesions gradually progress to multiple ground-glass opacities in the lungs, and some patients may have a dense consolidation in lobes. Interlobular septum in ground-glass opacities thickens, similar to "crazy-paving." In severe cases, patients may have diffuse lesions in both lungs, some of which show a "white lung" appearance. Air bronchograms and signs of blood vessel penetration are common. In the absorption or remission stage, ground-glass opacities and consolidation resolve without signs of "crazy-paving," and parenchymal subpleural fibrosis lesions may be observed. Thus, computed tomography can indicate the course of the disease and assess clinical severity and COVID-19 extent.
On some occasions, computed tomography routine use can lead to false positives, such as pneumonia cases that are not caused by SARS-CoV-2. However, the predictive ability of this diagnostic technique may improve over time. Also, patients are exposed to potential risks due to irradiation used during image acquisition. Another significant risk is due to cross-infection during the scanning process. For this reason, it is essential to have strict protective measures during a scan. Despite all these risks, computed tomography remains an important method for diagnosis, scope assessment, severity, and COVID-19 monitoring[66].
Lauer and collaborators (2020) estimated the duration of the COVID-19 incubation period. The study was conducted from a joint analysis of confirmed COVID-19 cases between January 4 and February 24 from reports and press releases from fifty provinces, regions, and countries, excluding the city of Wuhan, China[67]. The study took into account patient demographic characteristics, dates, and times of possible exposure, symptoms onset, fever onset, and hospitalization. Results showed 181 confirmed cases with identifiable exposure and onset identification of symptoms to estimate the COVID-19 incubation period. With these results, a median incubation period of 5 days was determined (within a 95% confidence interval, 4.5 to 5.8 days), and 97.5 percent of those infected who developed symptoms did so within 11.5 days after being infected (confidence interval, 8.2 to 15.6 days). Additionally, the study estimated that 101 out of every 10 000 cases would develop symptoms after fourteen days of monitoring or incubation quarantine for COVID-19.
Detection methods of SARS-CoV-2
The whole-genome sequencing of SARS-CoV-2 is one of the most comprehensive approaches for viral nucleic acid identification and quantification with high accuracy and reliability, making it possible to identify SARS-Cov-2 unequivocally. The first complete SARS-CoV-2 genome sequence available from GenBank has an accession number of MN908947. This sequencing allowed the identification of five typical open reading frames in the SARS-CoV-2 genome, ORF1ab polyprotein (7096 amino acids), protein S (1,273 amino acids), protein E (75 amino acids), protein M (222 amino acids), and protein N (419 amino acids). Subsequently, sequences of other SARS-CoV-2 genes have been reported using Illumina and Nanopore platforms[68]. Because whole-genome sequencing is relatively expensive, as it is complicated and requires long testing times, it is not suitable for large-scale and urgent testing. For this reason, alternative methods are required that allow results to be obtained quickly, relatively cheaply, and reliably.
Reverse transcription polymerase chain reaction is the most popular test method for SARS-CoV-2 detection that commonly uses samples from the back of a patient's nose or mouth. From these samples, ribonucleic acid is extracted and converted into its complementary deoxyribonucleic acid via a transcription reaction in reverse transcriptase enzyme presence. Later, specific gene fragments within complementary deoxyribonucleic acid are amplified using primers for specific targets. Primers are generally obtained from three regions of viral genome with conserved sequences:
- RdRP gene (ribonucleic acid -dependent gene that expresses ribonucleic acid polymerase) in open reading frame of ORF1ab region.
- Gene that expresses protein E.
- Gene that expresses protein N.
Assay can be designed as a two-target system, where one primer universally detects numerous coronaviruses, including SARS-CoV-2, and a second set of primers that only detects SARS-CoV-2[69].
Escutenaire and collaborators (2007) developed a reverse transcription polymerase chain reaction assay for generic coronavirus detection using ORF1b polyprotein and SYBR-Green 1 fluorophore as an initiator for detection[70]. The study evaluated human CoVs H-CoV-NL63, H-CoV-OC43, H-CoV-229E, and SARS-CoV. Results showed that this test is useful when there is a wide range of genetically distant viruses.
Liu and collaborators (2020) tested 4 880 patients from a hospital in Wuhan, China, for SARS-CoV-2 using a reverse transcription polymerase chain reaction technique. Study results showed that using bronchoalveolar lavage fluid samples and the ORF1ab polyprotein fragment genes as a target, a one-hundred percent detection rate of SARS-CoV-2 is achieved.
Kasteren and collaborators (2020) compared basic clinical and analytical performance of selected reverse transcription polymerase chain reaction kits from seven different manufacturers (Altona Diagnostics, BGI, CerTest Biotec, KH Medical, PrimerDesign, R Biopharm AG and Seegene)[71]. Information about kits can be founded on this website (accessed July 22, 2020). Study results showed a polymerase chain reaction efficiency of up to 96 percent for all assays and a LOD95 that varied within a six-fold range for various reverse transcription polymerase chain reaction kits targets.
On the other hand, the best known isothermal amplification methods are transcription-mediated amplification or self-sustained sequence replication, nucleic acid sequence-based amplification, signal-mediated amplification of ribonucleic acid technology, strand displacement amplification, rolling circle amplification, loop-mediated deoxyribonucleic acid isothermal amplification, isothermal multiple displacement amplification, helicase-dependent amplification, the isothermal amplification of single primer, and circular helicase-dependent amplification[72]. A more recently used isothermal nucleic acid amplification technique is based on short palindromic repeats grouped regularly between interleaved spaces, commonly known as CRISPR. These sequences can be recognized and cut by a bacterial enzyme set called clustered and regularly spaced short palindromic repeats-associated enzymes, exemplified by enzymes Cas12 and Cas13 families. These two enzyme families can be programmed to attack and cut viral ribonucleic acid sequences[73].
Lin and collaborators (2020) developed an assay based on loop-mediated isothermal amplification combined with reverse transcription that allows direct detection SARS-Cov-2 ribonucleic acid[74]. The authors called this assay iso-thermal LAMP-based method for COVID-19 (iLACO). This method showed similar performance with SARS-CoV-2 ribonucleic acid or cDNA samples. The iso-thermal LAMP-based method for COVID-19 assay is a low cost, fast, robust, simple, and accurate for SARS-CoV-2 detection.
Broughton and collaborators (2020) developed and performed clustered, and regularly spaced short palindromic repeats assay initial validation based on Cas12 enzyme for SARS-Cov-2 detection from ribonucleic acid samples[75]. The assay was called SARS-CoV-2 deoxyribonucleic acid (DNA) endonuclease-targeted short palindromic repeat transformer (CRISPR), or DETECTR. This assay performs reverse transcription and isothermal amplification simultaneously using loop-mediated amplification (Real-time loop-mediated deoxyribonucleic acid isothermal amplification)[76] of nasopharyngeal or oropharyngeal ribonucleic acid, followed by Cas12 detection of predefined coronavirus sequences. Finally, reporter molecule cleavage confirms virus detection. DETECTR assay based on clustered regularly interspaced short palindromic repeats is qualitative and only requires about forty-five minutes, with a positive predictive value of ninety-five percent and a negative predictive value of one hundred percent.
Another test types are serological tests characterized by blood analysis, serum or blood plasma, saliva, sputum, and other fluids where target molecules are found. These tests are based on antigen-antibody reactions that cause color changes through the use of conjugate linked to an enzyme and an enzyme substrate that serves to identify the presence and concentration of proteins[77]. These assays are called enzyme immunoassays or enzyme-linked immunosorbent assays (ELISA).
Antibodies generated by the host in response to viral proteins can be present for a long time, which reliably allows indirect detection of SARS-CoV-2. Antibody tests can be particularly helpful in tracking COVID-19. Antibody-based serological techniques can be qualitative or quantitative tests. The most commonly used qualitative tests consist of immunoglobulin A, immunoglobulin G, and immunoglobulin M detection.
Guo and collaborators (2020) developed a method for COVID-19 diagnosis based on evaluating humoral response against SARS-CoV-2, including immunoglobulin A, immunoglobulin M and immunoglobulin G response, using an enzyme-linked immunosorbent-based assay of recombinant viral nucleocapsid protein[78]. The study involved a bronchoalveolar lavage fluid sample. The study results demonstrated that a patient’s plasma with SARS-Cov-2 does not have cross-reactivity with another coronavirus, except SARS-CoV. An immunoglobulin M test showed high effectiveness for SARS-CoV-2 detection after fourteen (IQR, from 10 to 18) days from the onset of symptoms.
Infantino and collaborators (2020) used a specific chemiluminescent immunoassay for anti-SARS-Cov-2 Immunoglobulin M and Immunoglobulin G antibodies, called enzyme-linked immunosorbent assay and chemiluminescence immunoassay[79]. The assay was performed with Immunoglobulin M and Immunoglobulin G antibodies against SARS-CoV-2. The study results show that majority of COVID-19 patients studied have both Immunoglobulin M and Immunoglobulin G antibodies. The overall assay sensitivity was seventy-five percent. Immunoglobulin G antibodies manifest a specificity of one hundred percent. This makes the assay suitable for different immunity assessment protocols for patients.
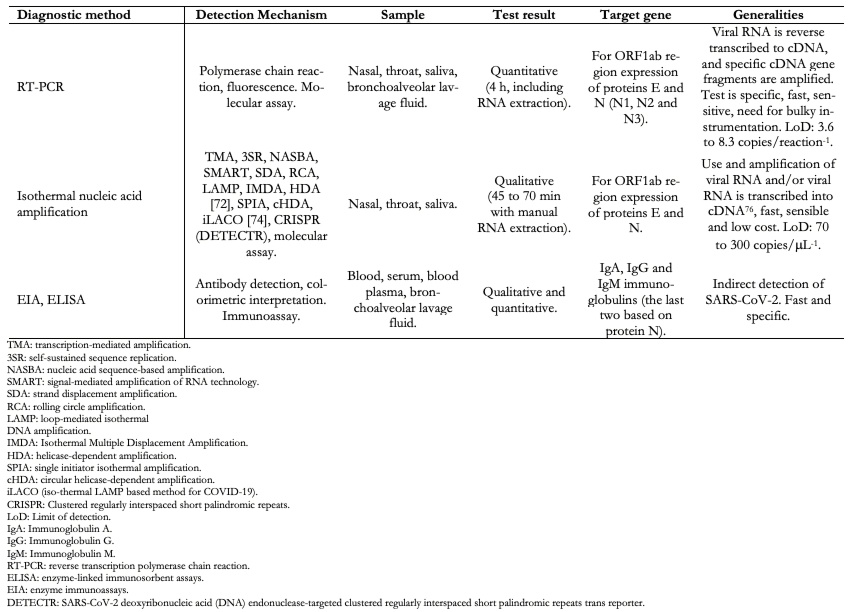
Table 4 shows the most representative COVID-19 diagnostic methods and some of their characteristics.
 Full size
Full size SARS-CoV-2 detection and monitoring should be carried out using two tests at different infection stages to ensure that patients infected are no longer virus carriers. The first test should be used to detect people infected with SARS-CoV-2 using serological tests[78],[79]. The second test should be used to detect the SARS-CoV-2 viral genetic material in a patient by reverse transcription polymerase chain reaction[70],[71] or an isothermal amplification of nucleic acid[74],[75].
Antivirals evaluated to combat SARS-CoV-2
Despite a global effort to develop drugs or vaccines to treat or prevent infection with SARS-CoV-2, there is currently no proven antiviral to control the COVID-19 outbreak or inactivate coronavirus. Therefore, the drugs used are not entirely effective, or published studies do not give certainty of their effectiveness. The current strategy for treating patients with COVID-19 also includes infection prevention and clinical support measures for severe patients with oxygen and mechanical ventilator support[80]. Wang and collaborators (2020) performed antiviral tests against SARS-CoV-2 using Vero E6 cells infected with SARS-CoV-2 at a multiplicity of infection of 0.05 at different remdesivir and chloroquine doses[81]. Vero E6 cells are African green monkey kidney epithelial cells belonging to a continuous, aneuploidic cell lineage. This means Vero E6 cells replicate through many replication cycles without aging with an abnormal chromosomes number and are used for vaccine production against viral diseases, while remdesivir is an antiviral prodrug that belongs to the nucleotide analogs group, which is incorporated into the nascent viral RNA chains.
This study results demonstrated a ninety percent maximal effective concentration of remdesivir against SARS-CoV-2 in Vero E6 cells was 1.76 micromolar. On the other hand, this study also evaluated chloroquine antiviral activity against SARS-CoV-2, the latter being a widely used antimalarial drug. Chloroquine block virus infection by increasing the endosomal pH required for virus/cell fusion. The study showed chloroquine acted in both the contagion phase and later. In addition to its antiviral activity, chloroquine has an immunomodulatory activity, which can synergistically increase its antiviral effect in vivo. After oral administration, chloroquine is widely distributed throughout the body, including the lungs. The ninety percent maximal effective concentration value of chloroquine against SARS-CoV-2 in Vero E6 cells was 6.90 micromolar.
It should be noted that the FDA recently authorized remdesivir for emergency use and chloroquine phosphate and hydroxychloroquine sulfate for the treatment of hospitalized patients with COVID-19.
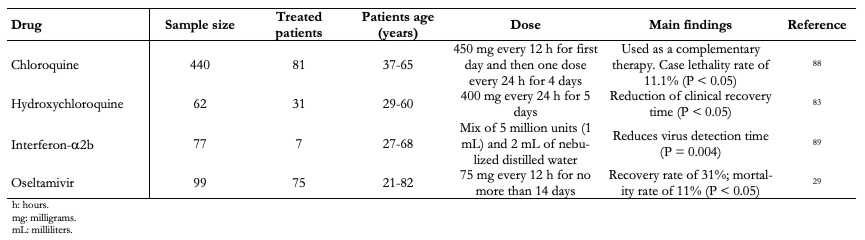
Furthermore, a small-scale French trial suggests hydroxychloroquine and antibiotic azithromycin help COVID-19 patients recover faster[82]. The patients received 600 milligrams of hydroxychloroquine daily, and their viral load on nasopharyngeal swabs was tested daily in a hospital. Depending on its clinical characteristics, azithromycin was added to the treatment. Untreated patients from another center and cases that rejected protocol were included as negative controls. Hydroxychloroquine treatment appeared to suppress the virus in fourteen of twenty people who received the drug for six days, compared with just two of sixteen people in the control group. Six people who received hydroxychloroquine along with azithromycin were reported "virologically cured" within six days. Despite the small sample size, this study suggests that hydroxychloroquine treatment is associated significantly with reduction-disappearance of viral load in COVID-19 patients, and its effect is enhanced by azithromycin. A more recent study, conducted by Chen and collaborators (2020) covered 62 patients, 31 treated with hydroxychloroquine, Table 5. The study results showed that 80.6 percent of treated patients showed notable improvement[83]. These results are promising; however, the implementation of randomized clinical trials with appropriate monitoring is required to determine if chloroquine and hydroxychloroquine prevent or effectively treat COVID-19[84].
Despite all potential positive effects that can be attributed to chloroquine and hydroxychloroquine and their wide distribution in the market at low costs, it is important to note these drugs have a very narrow margin between therapeutic and intoxication doses. Both drugs have been associated with cardiovascular disorders that can put the patient's life at risk. Therefore, chloroquine and hydroxychloroquine use should be subject to strict rules, and self-medication is not recommended[85]. It is also important to note that currently, double-blind, randomized studies are scarce, and consequently, the clinical efficacy of these drugs is unknown in COVID-19 treatment[86].
Another promising study for COVID-19 treatment was recently reported by Zhou and collaborators (2020). This study consisted of interferon-a2b use with or without umifenovir. This treatment reduced detectable virus duration upper respiratory tract and, in parallel, reduced elevated blood levels duration of inflammatory markers IL-6 and CRP with or without umifenovir. Interferon-a2b is a broad-spectrum antiviral that exhibits direct inhibitory effects on viral replication and reinforces an immune response to infection by a virus. The virus antiproliferation mechanism of interferon-a2b occurs through protein synthesis regulation and protein selective translation in inhibiting virus proliferation[87].
Table 5 lists clinical case series studies with some drugs that have involved a large number of patients and have shown promising results and will undoubtedly lead to trials with larger patient populations in the next future.
 Full size
Full size On the other hand, Richardson and collaborators (2020) used BenevolentAI, a great repository of structured medical information, including numerous connections drawn from scientific literature using artificial intelligence, along with a specific search for SARS-CoV-2, to identify approved antiviral drugs with the potential ability to block the viral infection process[90]. From this in silico study, Richardson and collaborators (2020) found baricitinib, a selective and reversible inhibitor of Janus kinases JAK1 and JAK2. Baricitinib is used to treat rheumatoid arthritis in adults. The study predicts that this drug reduces the virus's ability to infect lung cells with SARS-CoV-2.
Finally, in relation to the search for therapeutic agents and vaccines for human coronavirus, Liu and collaborators (2020) conducted an in-depth review of research papers and patents[91]. Patents reviewed highlight antiviral strategies involving small molecules and biological products, targeting complex molecular interactions in coronavirus infection and replication. Biologics include therapeutic antibodies, cytokines, nucleic acid-based therapies targeting virus gene expression, and various types of vaccines. Consequently, Liu and collaborators (2020) provide references for the continued development of human coronavirus-related vaccines and therapeutic agents.
In relation to this, Jackson and collaborators (2020) reported preliminary findings from the evaluation of a phase one vaccine to determine SARS-CoV-2 messenger ribonucleic acid vaccine safety and immunogenicity. Phase one involved 45 healthy participants, ages 18 to 55, who received a vaccine candidate in one of three dose levels (25, 100, or 250 micrograms) provided in two vaccinations 28 days apart[92]. The study results showed that after the first vaccination, antibody concentration measured on day 29 was higher at higher doses. After the second vaccination, antibodies measured on day 57 continued to increase with very good results with patients who received 250 micrograms doses, increasing antibodies amount by 5.6 times compared to the first vaccination. The study also managed to detect neutralizing activity after the second vaccination. There were some adverse effects of note in participants, including fatigue, chills, headache, myalgia, and pain at the injection site after the second vaccination.
Conclusions
SARS-CoV-2 spread to practically the entire world, causing severe damage to the health sector. The virus transmission routes are mainly from humans to humans; however, some recent studies indicate its possible transmission through air and water. The group of people at most risk by COVID-19 are adults over 60 years. People in this group with a chronic disease are highly susceptible to exhibit a COVID-19 severe state and require hospitalization.
SARS-CoV-2 diagnosis may be accompanied by chest computed tomography images to assess the grade of patient affectation. Furthermore, significant virus spread mitigation can be achieved by implementing a combined intervention based on quarantine, school closings, and workplace distancing. These preventive measures can reduce spread by approximately 78 percent. In addition to the above measures, this depends on the availability of reliable and cheap tests to monitor infected patients and those highly susceptible to contagion.
For SARS-CoV-2, detection reverse transcription polymerase chain reaction, iso-thermal LAMP based method for COVID-19, and DETECTR tests are recommended.
Regarding COVID-19 treatments, despite the existence of some study cases with chloroquine, hydroxychloroquine, remdesivir, interferon-a2b, and oseltamivir in hospitalized patients, it is still impossible to assert that there are treatments that efficiently and reliably control SARS-CoV-2. Randomized clinical trials are currently being conducted with the drugs mentioned above, and the real impact of these drugs will be known shortly.
Finally, it is worth mentioning that knowledge is being generated quickly and has free availability from practically all international publishing houses. Anything related to SARS-CoV-2 is allowed to advance at speed never seen before in COVID- 19 monitoring and control—a disease that shocked the whole world.
Notes
Authorship contributions
NFSS: prepared sections on coronaviruses classification, coronavirus virions structure and mechanism of SARS-CoV-2 action and origin of the β-coronavirus lineages in humans.
RSC: elaborated COVID-19 characteristics and symptoms, SARS-CoV-2 turned into a pandemic, detection methods of SARS-CoV-2 and SARS-CoV-2 antivirals. Abstract, introduction and conclusions sections were constructed jointly.
Acknowledgments
The authors thank the Universidad Tecnológica de la Mixteca for the facilities provided to develop this work and the reviewers for their recommendations that helped improve the manuscript.
Competing interest
The authors completed the ICMJE conflict of interest declaration and declared that they received no funds for realization of this article; do not have financial relationships with organizations that in last three years may have an interest in the article and have no other relationships or activities that may influence the content of this article. The forms can be requested by contacting the responsible author or the Editorial Committee of the Journal.
Funding
Universidad Tecnológica de la Mixteca funded the present work.

