Clinical reviews
← vista completaPublished on May 26, 2021 | http://doi.org/10.5867/medwave.2021.04.8198
What we know and don’t know on SARS-CoV-2 and COVID-19
Lo que sabemos y no sabemos sobre SARS-CoV-2 y COVID-19
Abstract
Coronavirus disease 2019 (COVID-19), caused by the SARS-CoV-2 virus discovered in December 2019 in Wuhan, China, has had an enormous impact on public health worldwide due to its rapid spread and pandemic behavior, challenges in its control and mitigation, and few therapeutic alternatives. In this review, we summarize the pathophysiological mechanisms, clinical presentation, and diagnostic techniques. In addition, the main lineages and the different strategies for disease prevention are reviewed, with emphasis on the development of vaccines and their different platforms. Finally, some of the currently available therapeutic strategies are summarized. Throughout the article, we point out the current knowns and unknowns at the time of writing this article.
Main messages
- In the course of a little over a year, SARS-CoV-2 and the disease it causes have spawned a massive worldwide research effort that has resulted in a large body of evidence, much of which has yet to be included in comprehensive narrative reviews.
- Updates on the pathology, clinical manifestations, prevention, and treatment are needed to keep up with the literature.
- Every one of these areas is frought with much uncertainty.
- Some aspects of COVID-19 diagnostics, treatment, and prevention have already achieved consensus based on novel research.
- Vaccines are an exceptionally dynamic field of great interest to public health.
- The present review is not systematic but strives to provide comprehensive guidance on the state of the art as we know it to this date.
Introduction
On December 31, 2019, the World Health Organization (WHO) reported a series of pneumonia cases caused by an unknown agent in Wuhan City, Hubei Province, China. In January 2020, the cause of this infection was reported to be a novel coronavirus, initially named 2019-nCoV (novel coronavirus 2019). WHO subsequently named SARS-CoV-2 and the disease caused by it COVID-19 (coronavirus disease 2019). The infection spread rapidly through China and its neighboring countries, spreading throughout the world in a few weeks, probably facilitated by inter-city travel and tourist arrivals in the context of the Lunar New Year being celebrated in China at that time. The global repercussions of the epidemic were quickly evident, which is why on January 30, 2020, WHO declared COVID-19 a public health emergency of international concern and in March of the same year declared it a pandemic[1],[2],[3],[4].
Since the publication of the first cases reported in Wuhan, China, in December 2019[5], the disease caused by SARS-CoV-2 (COVID-19) has resulted in more than 138 million cases and 3 million deaths worldwide to date[6], and is one of the most devastating pandemics of recent times.
This article will review the main epidemiological and clinical features of the disease caused by SARS-CoV-2 and some virological, diagnostic, and therapeutic aspects.
Pathogenesis
Coronaviruses are members of the subfamily Coronavirinae in the family Coronaviridae and the order Nidovirales. This subfamily consists of four genera: alphacoronavirus, betacoronavirus, gammacoronavirus, and deltacoronavirus, based on their phylogenetic relationships and genomic structures. Alphacoronaviruses and betacoronaviruses infect only mammals, whereas gammacoronaviruses and deltacoronaviruses infect birds, but some of them can also infect mammals. Coronaviruses are large, enveloped, single-stranded RNA viruses found in humans and other mammals, such as dogs, cats, chickens, cows, pigs, and birds, and can cause respiratory, gastrointestinal, and neurological diseases. The most common coronaviruses affecting humans are 229E, OC43, NL63, and HKU1, which usually cause common cold symptoms in immunocompetent individuals. In 2002, an outbreak of a new coronavirus was described in the Guandong province of China called SARS (Severe Acute Respiratory Syndrome). Ten years later, in 2012, another highly pathogenic coronavirus, MERS-CoV (Middle East Respiratory Syndrome Coronavirus), affected several countries in the Middle East. We are currently experiencing a third coronavirus outbreak, this time due to SARS-CoV-2, a virus that shares 79% of the genomic sequence with SARS and 50% with MERS, and with pandemic behavior. SARS was transmitted to humans from civets, MERS from dromedary camels, and SARS-CoV-2 from pangolins. All three viruses are thought to have originated in bats[7],[8],[9]. Domestic animals may play important roles as intermediate hosts that allow transmission of the virus from natural hosts to humans, supporting the theory that the virus originated in the Wuhan market. However, this has been disputed given the finding in France of SARS-CoV-2 by PCR in a stored sample from a patient who had pneumonia in late 2019, suggesting that the virus may have been circulating earlier than currently believed[10].
SARS-CoV-2 has a diameter of 60 nm to 140 nm and distinctive spikes, ranging from 9 nm to 12 nm, giving the virions the appearance of a solar corona. Coronaviruses can adapt and infect new hosts through recombination and genetic variation[8], which has led to the emergence of several variants of worldwide interest, the most recognized to date being B.1.1.7 (UK), B.1.351 (South Africa), B.1.1.28 (Brazil), B.1.427/B.1.429 (USA) and, more recently, B.1.617 (India)[11],[12].
SARS coronaviruses use angiotensin-converting enzyme 2 (ACE2) as a receptor and invade mainly bronchial ciliated epithelial cells and type II pneumocytes. On the other hand, MERS-CoV uses dipeptidyl peptidase-4 (DPP4) as a receptor and invades non-ciliated bronchial epithelium and type II pneumocytes[9].
Once the virus comes into contact with the respiratory mucosa, the SARS-CoV-2 spike glycoprotein (S) binds to surface receptors on the host cells and mediates viral entry by interacting with the angiotensin-converting enzyme 2 receptors (ACE2), resulting in disease transmission and pathogenesis. The transmembrane protease serine 2 (TMPRSS2) in the host cell further promotes viral uptake by cleaving ACE2 and activating the SARS-CoV-2 S protein. Upon binding to respiratory tract epithelial cells, SARS-CoV-2 begins to replicate and migrate into the airways and enters the alveolar epithelial cells of the lungs. Rapid replication of SARS-CoV-2 in the lungs can trigger a strong immune response, mediated by inflammatory signaling molecules, T lymphocytes, monocytes and neutrophils released from infected cells and alveolar macrophages. Cytokine storm syndrome causes acute respiratory distress syndrome and respiratory failure, which is considered the leading cause of death in patients with COVID-19[7],[8],[13],[14].
| What we know | What we don't know |
|
|
Clinical manifestations
The disease caused by SARS-CoV-2, COVID-19, is characterized mainly by respiratory involvement with various signs and symptoms that have been described as the pandemic unfolded.
The viral incubation period varies between 2 to 14 days, with an average of 4 days[15],[16], while viral shedding varies by clinical severity. It has been shown that in mild to moderate cases, the highest risk of transmission or contagion begins two days before the onset of symptoms until approximately five days after the onset of symptoms. However, viable viruses have been found in cell cultures up to 7 to 10 days after the onset of symptoms[17]. This period can last up to 20 days in severe cases and immunocompromised patients[18]. It is important to note that the finding of viral RNA by polymerase chain reaction (PCR) does not equate to a viable virus with infecting capacity being present and can be found in significant quantities up to several months after illness[18].
The most frequently reported symptoms are cough, myalgia, headache, and fever, but dyspnea, odynophagia, diarrhea, nausea and vomiting, anosmia, ageusia, nasal congestion, fatigue, and chest pain have also been reported[15],[19],[20],[21].
It is estimated that one-third of SARS-CoV-2 infections are asymptomatic[22], although this is difficult to determine since a percentage of patients considered asymptomatic at the time of confirming the diagnosis with a positive PCR test may present symptoms during the following days. This group of patients may have other manifestations of the disease, such as laboratory alterations or abnormalities on chest CT scan[23]. These individuals can infect others and may not isolate due to the lack of symptoms that would warn of the disease in time[24],[25].
The disease can have a mild (80%), severe (15%), or critical (5%) course, with an estimated case fatality rate of 2.3% in the first reports[26]. Mild disease corresponds to the absence of pneumonia or mild pneumonia; severe disease corresponds to dyspnea, hypoxia, or involvement of more than 50% of the lung parenchyma on chest imaging; and critical disease is characterized by respiratory failure, shock, or multiorgan dysfunction[26],[27].
The recovery time of the disease is variable, depending on various factors such as the severity of the condition, age, and comorbidities. Most mild cases recover within the first two weeks of illness; however, prolonged manifestations of COVID-19 of variable duration averaging three months have been described. This form of presentation is most frequently seen in people who have had a severe or critical illness, and the most frequent clinical manifestations are fatigue (53%), dyspnea (43%), joint pain (27%), and chest pain (22%)[28],[29]. Neuropsychiatric symptoms such as anxiety disorder, dementia, and insomnia have also been described[30]. It is estimated that over 80% of patients maintain at least one symptom 60 days after overcoming acute infection[28].
Although severe cases can occur at any age, according to published data[26],[31],[32],[33],[34],[35],[36],[37],[38], older age, male sex, obesity, cardiovascular morbidities such as hypertension, diabetes, dyslipidemia, and coronary heart disease; chronic obstructive pulmonary disease and other lung conditions; chronic kidney disease, cancer, smoking, and a history of solid organ and hematopoietic precursors transplantation; have been associated with a higher risk of developing severe COVID and even higher mortality; have been associated with an increased risk of developing severe COVID and increased mortality. Another factor that has been associated with increased risk of COVID-19 complications is inequity in the social determinants of health, including socioeconomic status[39] and belonging to underrepresented racial/ethnic groups[40], among others.
Transmission
As the pandemic progresses, a deeper understanding of the transmission mechanisms has been achieved, starting with the zoonosis theory from the Wuhan animal market to the current evidence supporting person-to-person transmission, which is now considered the main route of transmission of the disease[41].
The primary mode of transmission is through close contact (less than 1.5 to 2 meters distance) by inhaling air with respiratory particles from the respiratory tract of a SARS-CoV-2 infected person. The large droplets produced by talking, coughing, or sneezing can contact the respiratory mucosa of a susceptible host, invade it, and cause disease. Infection can also occur if these respiratory secretions contaminate a person’s hands (e.g., by touching contaminated surfaces) and then touching the eyes, nose, or mouth. Nonetheless, more recent evidence suggests contact with contaminated surfaces is not a significant route of transmission[41],[42]. Therefore, maintaining a physical distance of at least 1.5 meters between people, the widespread use of masks and timely and frequent handwashing have become the main measures to prevent the transmission and spread of SARS-CoV-2[43]. Another transmission route is aerosols—particles smaller than droplets that remain suspended in the air for a longer time and travel longer distances. Recently, increasing evidence has been published regarding this transmission route in the spread of the pandemic, making more evident the role of indoor ventilation as a fundamental measure to reduce the risk of transmission[43],[44],[45]. Although SARS-CoV-2 has been detected in non-respiratory samples (stool, blood, semen, urine), it has not been shown to play a significant role in viral transmission[18],[46].
Variants
As the pandemic has progressed, different variants have been identified as a result of genetic mutations, a common process inherent to viruses, which is exacerbated by the high rates of community transmission worldwide (> 100 new cases of COVID-19 per million population per day)[47]. Compared to other viruses, such as influenza and HIV, SARS-CoV-2 has a lower mutation rate, and most mutations do not impact viral function.
To date, five variants of SARS-CoV-2 have generated interest worldwide because of the potential impact on transmission dynamics and disease severity[11],[12],[48],[49]. These variants, termed “variants of concern,” have in common an amino acid substitution in the spike protein (D614G, glycine for aspartic acid), which translates into increased replication and transmission capacity, explained by a higher viral load in the upper respiratory tract and stronger affinity for the ACE2 receptor. Data are still lacking to affirm that these variants increase the risk of hospitalization by COVID-19 or affect the neutralizing capacity of anti-spike antibodies[50],[51],[52],[53], but some studies have shown that some of these variants of interest are associated with a higher rate of complications[54],[55] and lower susceptibility to the immune response generated by some vaccines[56],[57],[58] (Table 1).
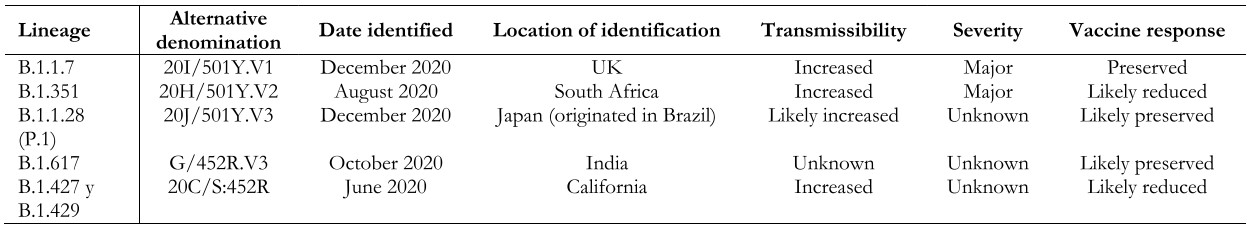 Full size
Full size Lineage B.1.1.7 (a.k.a. 20I/501Y.V1 VOC 202012/01): Identified in December 2020 in the United Kingdom. Retrospectively, it was found that circulation began in September 2020, so its dissemination to other countries during that period is unknown. Variant B.1.1.7 has a mutation in the receptor-binding region of protein S. Some studies suggest that this new variant is significantly more transmissible than others, with an estimated potential increase in the reproductive number of 0.4 or more and with estimated increased transmissibility of up to 90%[59]. It has also been observed to increase the mortality risk, although the research is still unfolding[54],[55]. Variant B.1.1.7 is associated with a slight decrease in neutralizing antibody titers but above the levels associated with protection, with a low risk of reinfection[60]. Regarding the impact on vaccine-generated immunity, this variant does not appear to significantly reduce the efficacy of the Sinovac, Pfizer/BioNTech, Moderna, Novavax, and AstraZeneca vaccines[59]. To date, B.1.1.7 has been identified in 137 countries according to the WHO epidemiological report of April 20, 2021[49].
Lineage B.1.351 (a.k.a. 20H/501Y.V2): identified in South Africa in August 2020 and reported in 85 countries[49]. This variant has multiple mutations in the spike protein, several of them in common with variant B.1.1.7. There is now evidence that the response to neutralizing antibodies and vaccines may be diminished compared to other variants[61],[62] because of a specific mutation in the spike protein (E484K), although the evidence is still unfolding[63],[64],[65]. It has also been described that mutations present in this variant may help the virus evade the immune response triggered by previous SARS-CoV-2 infection, with a consequent risk of reinfection. This variant presents a higher risk of transmission, with the possibility of generating a more severe clinical picture compared to the original variant[66].
Lineage B.1.1.28 (a.k.a. 20J/501Y.V3 or P.1): first identified in December 2020 in Japan in travelers’ samples from Brazil. This variant contains three mutations in the receptor-binding domain of the spike protein. So far, there is no clear evidence regarding the impact of this mutation on the transmissibility and aggressiveness of the disease. Cases of reinfection with this variant and a reduction in the neutralizing capacity of antibodies have been reported, with the possibility of presenting reinfections or, eventually, lower response to vaccines[57]. However, there is also evidence supporting the efficacy of at least one vaccine (Sinovac) in a high-circulation environment of this lineage[67],[68],[69]. According to the WHO epidemiological report, as of April 20, 2021, this variant had been reported in 52 countries[49].
Lineage B.1.627 (a.k.a. G/452R.V3): first detected in India in October 2020. It has gained special interest given the rapid increase of COVID-19 cases in India with a significant epidemiological impact in this country, even though there is no evidence regarding the involvement of this variant in transmission or morbidity and mortality associated with the infection. The limited evidence currently available has not shown a decrease in the effectiveness of vaccines against this variant. To date, the presence of this lineage has been reported in 21 countries[70].
B.1.427 and B.1.429 (a.k.a. 20C/S:452R) lineage: identified in California (USA), structurally similar to each other. They are associated with increased transmissibility and, apparently, reduced neutralization capacity[56]. These lineages have been listed as “variants of concern” by the US Centers for Disease Control and Prevention (CDC), but not by the World Health Organization[12].
In addition, other variants called “variants of interest” have been reported, with lesser but significant clinical impact than the “variants of concern.” Outstanding in this group are the B.1.526 and B.1.526.1 lineages (New York), B.1.525 (United Kingdom/Nigeria), P.2 (Brazil), among others[71].
The importance of these variants regarding transmissibility, severity, antibody neutralization capabilities, and potential impact on the effectiveness of COVID-19 vaccines is still under investigation. The detection of other mutations potentially impacting public health is also evaluated and monitored through surveillance and genetic sequencing in different countries worldwide[11],[47],[48],[49].
Reinfection
The possibility of reinfection with all four known human seasonal coronavirus infections, even in the presence of pre-existing antibodies, is not unusual; however, reactivated, relapsing, or latent infection appears less likely and has not been described for the coronavirus family[72]. Isolated cases of reinfection with SARS-CoV-2 have been documented in individuals with a history of previous COVID[72],[73], estimating a 0.02% risk with a 0.36 incidence rate[74]. Establishing the diagnosis of reinfection is challenging, and molecular testing alone is not of great utility due to the possibility of prolonged respiratory excretion of viral RNA after acute infection. A second positive PCR test in a patient who has recovered from COVID-19 does not necessarily indicate reinfection. Factors that increase the likelihood of reinfection include a longer time interval since the first infection, a high level of viral RNA in the repeat test, and undetectable IgG antibody at the time reinfection is considered. However, reinfection can only be confirmed by genomic sequencing to establish that the infections were caused by two different viruses[72],[73],[74].
| What we know | What we don't know |
|
|
Diagnostics
Molecular testing
The first genetic sequencing of SARS-CoV-2 was performed in January 2020, only a few weeks after the first cases were reported, making a molecular test for diagnosis available early in the course of the pandemic[9],[75]. Detection of SARS-CoV-2 RNA based on reverse transcription-polymerase chain reaction (RT-PCR) from respiratory samples is the standard for diagnosis; however, the sensitivity of the tests varies with the timing of the test with exposure. Thus, the test’s sensitivity can range from 30% in the first four days post-exposure to 80% three days after symptom onset. Factors associated with higher sensitivity of the results include correct specimen collection technique, longer time since exposure, and the point of origin of the specimen (lower airway specimens, such as those obtained by bronchoalveolar lavage, which is more sensitive than upper airway specimens). Recently, techniques have been developed from saliva samples, which may be an alternative sample source requiring less personal protective equipment and fewer supplies. It is also possible to detect SARS-CoV 2 in stool[8].
Antigen testing
Another diagnostic test for COVID-19 is antigen testing, which can be performed rapidly and at point-of-care and, therefore, may be more accessible with a faster turnaround time to results than molecular tests, although it is less sensitive than molecular tests. Antigen testing can be useful in certain situations, provided that the possibility of false negatives is considered and results are interpreted based on pre-test probability. Frequently, the result of an antigen test must be confirmed by molecular testing (PCR)[76],[77].
Serological tests
Various serologic tests have been developed to support the diagnosis of disease and the assessment of vaccine response; however, the presence of antibodies may not confer immunity because not all antibodies produced in response to infection are neutralizing. It is not known whether the presence of antibodies changes susceptibility to subsequent infections or how long antibody protection lasts.
Different tests for anti-SARS-CoV-2 antibodies are commercially and experimentally available, using different technologies to qualitatively or quantitatively measure individual immunoglobulins (IgM, IgG, or IgA) or total antibodies (predominantly IgM, IgG, but also including other antigen-specific immunoglobulins)[78]. These serological tests detect SARS-CoV-2 antigens, specifically spike protein (S) or nucleocapsid (N). IgM antibodies are detectable within five days of infection, with the highest levels during weeks two to three of illness. In contrast, the elevation of IgG antibody titers is seen approximately 14 days after symptom onset, although, in some patients, both immunoglobulins are elevated simultaneously[78]. Secretory IgA is central to mucosal immunity, but its kinetics have not yet been elucidated, so its measurement is uncommon. Serologic assays include point-of-care assays and high-throughput enzyme immunoassays; however, test performance, accuracy, and validity are variable[8]. In general, tests that measure IgM have lower sensitivity for detecting past infection than those that detect IgG or total antibody; and those that detect IgA tend to have lower specificity[78].
Specificity is important in seroprevalence studies when the community prevalence of past infection is expected to be low. Serological tests must have high sensitivity and specificity (≥ 99.5%) to be valid[78]. Detection of IgG or total antibody at 3 to 4 weeks after symptom onset provides the highest sensitivity and, therefore, the lowest rate of false-negative results compared with other immunoglobulin classes. IgG or total antibody tests also provide high specificity and reduce the rate of false-positive results compared with other antibody types[78]. Unlike other viral infections where IgM tests show high sensitivity soon after symptom onset compared to IgG, the sensitivity of IgM against SARS-CoV-2 is relatively low initially, and there is no significant increase over time as seen with IgG or total antibodies. The use of IgM testing alone could result in increased false-negative rates compared to IgG or total antibody testing[78].
The most commonly used laboratory platforms for serological diagnosis of SARS-CoV-2 are lateral flow, enzyme-linked immunosorbent assay (ELISA), and chemiluminescence or electrochemiluminescence immunoassay (CLIA or ECLIA). Lateral flow tests require a drop of blood, serum or plasma applied to a test strip and provide rapid results (15 to 30 minutes), making it an optimal test for point-of-care diagnosis and large, population-based seroprevalence studies. ELISA and CLIA/ECLIA assays are more complex laboratory techniques than lateral flow, useful for high-throughput testing using serum, plasma, or potentially dry blood, and allow quantifying binding antibody titers and determination of neutralizing antibodies[78]. It is important to mention that different vaccines use different antigens to stimulate the immune response, so if an individual receives a spike protein-based vaccine, they will generate anti-S-protein antibodies, for example, and not necessarily against other viral antigens. In addition, there are different components of the immune response, such as the humoral response and the cellular response, and the magnitude of specific antibody production may vary between individuals[79],[80],[81],[82]. Consequently, it is not appropriate to evaluate the immune response to vaccines exclusively based on serological tests, but it is essential to determine efficacy and effectiveness using clinical outcomes[83],[84].
Implications of the humoral and cellular immune response
Multiple studies have been conducted worldwide to determine the prevalence of individuals with or without symptoms of COVID-19 and with or without a confirmatory microbiological diagnosis who show evidence of having developed antibodies to SARS-CoV-2. To date, there is no clarity regarding the relationship between severity of infection, antibody titers, and risk of reinfection among different individuals and the absence of seroconversion in some individuals nor the role of cell-mediated immunity in the immune response to COVID-19[74],[79],[81],[82],[85],[86],[87]. Neutralizing antibodies are detected in approximately 40% to 70% of infected individuals; at least 30% of patients have no detectable antibody levels, and less than 15% achieve high neutralizing titers in vitro[87]. An association between neutralizing antibody titer and severity of COVID-19 disease has been observed, and those who have mild symptoms or are asymptomatic are less prone to generate a neutralizing response[87].
Seroprevalence studies
Worldwide, reported seroprevalence varies by factors such as the population studied (e.g., health care workers vs. general population) and the impact of the pandemic in the setting where the study is conducted (the evolution of the pandemic and the incidence rate vary according to the country, the season of the year, policies implemented, among other factors). Results range from prevalences as low as 0.4% to rates of 22%[88],[89],[90]. Publicly available software tools such as the Canadian Serotracker platform[91] will be very useful to assess the evolution and impact of the pandemic worldwide.
| What we know | What we don't know |
|
|
Treatment
While the management of COVID-19 is mainly based on symptomatic relief in mild to moderate cases and on ventilatory and, ultimately, hemodynamic support in severe or critical cases, the following section will describe some pharmacological interventions that have been shown to have an impact on the morbidity and mortality caused by COVID-19.
Corticosteroids
In patients with severe COVID, requiring supplemental oxygen or ventilatory support, the use of glucocorticoids, particularly dexamethasone, hydrocortisone, and methylprednisolone, has been shown to decrease 28-day mortality, as well as reduce the likelihood of requiring mechanical ventilation[92],[93],[94],[95],[96],[97],[98]. Particular caution should be taken with the adverse effects associated with the use of corticosteroids, such as hyperglycemia and bacterial or fungal superinfections[94],[99]. On the other hand, the use of inhaled budesonide has shown promising results in recent studies, accelerating recovery in early-stage cases of COVID-19 and decreasing the need for hospitalization or emergency department consultation[100],[101].
Remdesivir
The use of remdesivir—a nucleoside analog polymerase inhibitor—has been shown to decrease recovery time in adults hospitalized for COVID-19[102], as well as the need for mechanical ventilation, with no significant impact on mortality, when used in the first week of the disease[98],[99],[103],[104]. This benefit is more significant when remdesivir is associated with baricitinib[105],[106] (see below).
Interleukin-6 inhibitors
Elevation of pro-inflammatory interleukins, including IL-6, is associated with unfavorable outcomes and increased mortality in SARS-CoV-2, so their inhibition and the blockade of the inflammatory pathways may prevent disease progression[107]. Sarilumab and tocilizumab are monoclonal antibodies that neutralize the IL-6 receptor, while siltuximab is a direct inhibitor of this interleukin[108]. Some studies have shown that they could decrease the need for mechanical ventilation, the length of intensive care stay, hospitalization[99], and mortality[109],[110].
JAK inhibitors
Janus kinase 1 and 2 inhibitors are drugs that interfere with intracellular signaling of some interleukins (IL-2, 6, and 10), granulocyte and macrophage colony-stimulating factors, and interferon gamma, blocking the process of viral endocytosis by inhibiting AP2-associated protein kinase 1[105],[106]. Baricitinib and ruxolitinib have been shown to have some impact on reducing mortality, the need for mechanical ventilation, and the duration of hospitalization for COVID-19[99]. The association of remdesivir with baricitinib has been shown to reduce recovery time and accelerate clinical improvement in patients with COVID-19, especially in those with the need for high-flow oxygen or noninvasive ventilation[105],[106].
Colchicine
The use of colchicine in patients with mild to moderate disease—those who do not require hospitalization—could decrease mortality, the need for mechanical ventilation, and the duration of hospitalization for COVID-19. Nonetheless, there are ongoing studies that will provide further evidence for its usefulness in the management of this disease[98],[99],[111].
Interventions that have not consistently demonstrated usefulness in the management of COVID-19
The evidence available to date has determined that the risk of using some drugs outweighs the benefit in the treatment of COVID-19, including chloroquine, hydroxychloroquine, azithromycin, ritonavir-boosted lopinavir, favipiravir, and ivermectin[98],[99],[106],[112],[113],[114],[115],[116].
Recently, an open-label randomized study with a large sample size showed that the use of convalescent plasma has no significant impact on survival or other outcomes[117].
Empirical antibiotic treatment is not recommended in patients with COVID-19, regardless of the severity of the condition, unless there is significant evidence of bacterial infection (compatible clinical picture associated with the appearance of new infiltrates on chest imaging and positive cultures). It has been reported that more than half of patients with COVID-19 are prescribed antimicrobial therapy during the course of the disease, with bacterial infection confirmed in only 3.5% of cases[118],[119]. If antibiotic therapy is given, the indication should be re-evaluated daily, abbreviated courses of antimicrobials should be sought and adjusted to culture results[120].
| What we know | What we don't know |
|
|
Prevention
Personal prevention measures
Considering that the main route of transmission of SARS-CoV-2 is through droplets and aerosols generated from respiratory secretions, the interventions most proven to prevent infection are the universal use of masks, frequent and timely handwashing, physical distancing between people, ventilation of enclosed spaces, and avoidance of non-essential gatherings indoors and in crowded outdoor spaces[121].
Vaccines
Thanks to the global interest in controlling the pandemic, different vaccine platforms have been developed rapidly. When writing this review, 93 vaccines are in clinical development and 184 in the preclinical stage, as reported by the World Health Organization; 25 are in phase 3[122].
There are different technological platforms for SARS-CoV-2 vaccines, of which the most widely used are shown in Table 2[123],[124],[125].
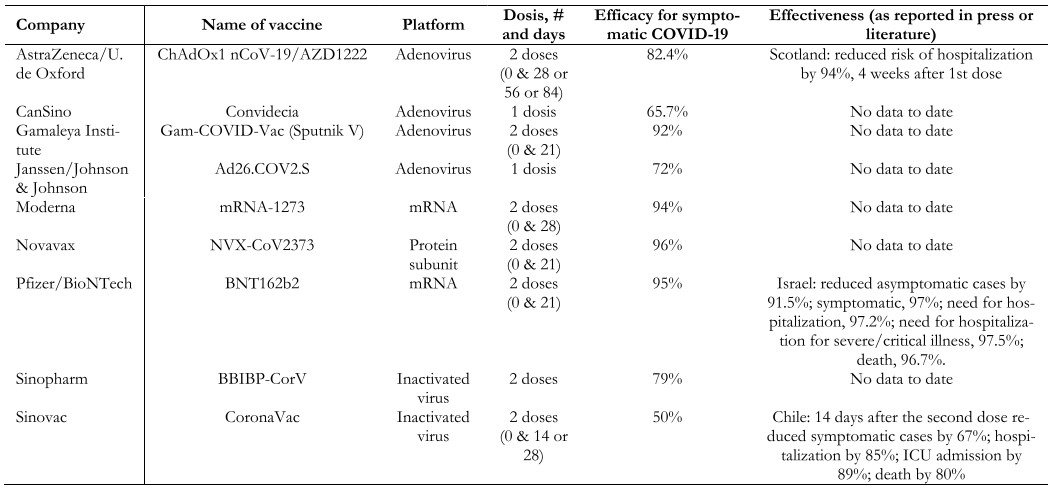 Full size
Full size Live attenuated virus. The first vaccine of this type to be developed was the smallpox vaccine (1978). These vaccines are produced by generating a genetically weakened version of the virus, maintaining a limited replication capacity without producing disease, but inducing an immune response similar to that generated by natural infection. The immune response generated by this type of vaccine targets both structural and non-structural viral proteins through cellular and antibody-mediated immune responses. The disadvantage of these vaccines is safety in immunocompromised patients and the difficulty involved in modifying the virus. Examples of live virus vaccines are measles, mumps, yellow fever, and shingles. Few SARS-CoV-2 vaccines use this platform, and none of them are in phase 3 clinical trials.
Inactivated virus. The first vaccine based on an inactivated virus was typhoid in 1986. These vaccines are developed using viruses treated with chemicals (e.g., formaldehyde), heat, or radiation, thus canceling their replicative capacity but maintaining their ability to generate an immune response. When the complete virus is presented to the immune system, in addition to generating a response against the spike protein of SARS-CoV-2, it also does so against the matrix, the envelope, and the nucleoprotein of the virus. Large quantities of the virus with infectious capacity grown in cell culture are required to produce this type of vaccine. Examples are hepatitis A, polio, influenza, pertussis, and rabies. Among the vaccines against COVID-19 that use this platform are Sinovac and Sinopharm.
DNA/RNA-based. This is a platform developed in recent years, and the SARS-CoV-2 vaccine is the first of this type to be used in humans outside the experimental setting. These vaccines consist of DNA or RNA fragments that code for a target antigen, which in the case of SARS-CoV-2 is often the spike (S) protein, allowing the vaccine recipient to express these antigens and then induce a humoral and cellular immune response against them. An advantage of these vaccines is the ease and speed of large-scale production; however, their disadvantage is the need to store and maintain them at low temperatures (between -20 and -70ºC), because of the significant logistical problem for their distribution, especially to remote and difficult-to-access locations. The most widely used COVID-19 vaccines worldwide with this platform are those of the Pfizer and Moderna laboratories.
Protein subunit-based vaccines. The first vaccine of this type to be used was the anthrax vaccine in 1970. They are based on purified virus particles or protein antigens. Research is being conducted on vaccines with the spike (S) protein and the RBD (receptor binding domain) protein. An advantage of these vaccines is that they can be produced without manipulating the live virus. Examples of this type of vaccine are hepatitis B, pneumococcus, and meningococcus, among others. It is currently the most widely used platform in clinical studies (31%)[122].
Vector vaccines. This is a relatively new technology, first used on a large scale for the Ebola virus in 2019. Vector vaccines use a genetically modified virus capable of producing proteins that induce an immune response in the vaccine recipient, such as the spike (S) protein of SARS-CoV-2. To prevent the recipient’s immune system from responding against this viral vector before the proteins needed to mount the immune response against the disease to be prevented are produced, viruses that do not affect humans, such as the chimpanzee adenovirus used in the production of the AstraZeneca laboratory’s vaccine, can be used. Other vaccines using this type of platform include those from CanSino, Janssen, and Gamaleya.
Over 600 million people worldwide have received at least one dose of the COVID-19 vaccine[126]. The duration of immunity generated by the vaccines and whether there are significant differences between the different platforms is yet unknown. We still do not know to what extent the genetic variants of the SARS-CoV-2 virus will affect the secondary immune response to infection or vaccination, nor how the genetic and environmental aspects specific to each patient will determine the immune response.
Data on the real-world effectiveness of some of the vaccines being used are now increasingly available. In Israel, the Pfizer/BioNTech vaccine showed significant concordance with published efficacy in the phase 3 study with very high effectiveness despite the prevalence of the B.1.1.7 (UK) variant at an estimated 94.5% (Table 2)[127]. An unpublished study conducted by the Ministry of Health of Chile showed that 14 days after the second dose, the CoronaVac vaccine from the Sinovac laboratory was 67% effective in preventing symptomatic COVID-19, 85% in preventing hospitalizations, 89% in preventing admission to the intensive care unit and 80% in preventing death from COVID[69]. Another study conducted by the CDC in the United States showed that the Pfizer and Moderna vaccines reduced the risk of infection by 90% two or more weeks after administering the second dose and by 80% after the first dose[128].
| What we know | What we don't know |
|
|
Conclusions
SARS-CoV-2 and the disease it produces, COVID-19, has generated an unprecedented global health impact and is likely to be a global public health problem for a long time to come. Different questions have been raised regarding the evolution of the pandemic as part of the world’s population is undergoing vaccination, there is no clarity on the duration of the immunity generated by both vaccines and natural infection, nor if collective immunity will be effectively achieved by reaching a significant coverage of the population or if it will be necessary to vaccinate again after a while as occurs with other respiratory viruses such as influenza. Likewise, there is little clarity regarding the impact of the different genetic variants on the post-infection immune response and the possibility of reinfection. Nor do we know what the level of the immune response to the different COVID vaccine platforms that have been developed will be in the long run. Different diagnostic and therapeutic strategies have been rapidly developed during this little more than a year of pandemic, while the scientific, academic, and policy worlds continue striving to increase the knowledge and technological developments necessary to combat this infection.

