Estudios originales
← vista completaPublicado el 9 de enero de 2026 | http://doi.org/10.5867/medwave.2026.01.3049
Identificación y perfil de susceptibilidad de microorganismos aislados directamente de orina en un estudio transversal usando espectrometría de masas por desorción/ionización láser asistida por matriz con tiempo de vuelo y Phoenix M50
Identification and susceptibility profile of microorganisms isolated directly from urine in a cross-sectional study using matrix-assisted laser desorption/ionization time-of-flight and Phoenix M50
Abstract
Introduction This study evaluates the efficacy of matrix-assisted laser desorption ionization–time of flight technology and the Phoenix™ M50 system for bacterial identification and antimicrobial susceptibility testing from bacterial concentrate obtained from urine samples, reducing diagnostic time to 24 hours compared to the traditional 72 hours in a public hospital in Antofagasta, Chile.
Methods Through differential centrifugation, a bacterial concentrate is obtained directly from urine, allowing the preparation of a McFarland standard for identification and susceptibility studies. We compared the identification and minimal inhibitory concentration results obtained from the bacterial concentrate with those obtained from the strain isolated in culture.
Results 380 samples were analyzed after exclusions. Direct identification showed 93.4% sensitivity and 73.8% specificity, with moderate agreement (κ=0.604) versus culture. had 98.5% concordance. For antimicrobial susceptibility test, Phoenix M50 performed well with , meeting Cumitech 31A standards for 13 out of 19 antibiotics. In contrast, only 8 of 19 antibiotics met the criteria for .
Conclusions The antimicrobial susceptibility method varies depending on the species-antibiotic combination; therefore, specific studies for each species are crucial. Despite these challenges, the direct method offers significant advantages in diagnostic speed and emphasize its potential for improving clinical decision-making, though further validation and protocol refinement are needed, particularly for Gram-positive pathogens.
Main messages
- This study addresses the need to reduce the diagnostic time for urinary tract infections using technologies such as matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) and the Phoenix™ 50 system.
- Despite advancements in Gram-negative bacteria, the method showed limitations in the identification of Gram-positive bacteria.
- The main finding was the high accuracy in identifying E. coli and K. pneumoniae, along with a significant improvement in diagnostic speed, potentially reducing the turnaround time from 72 to 24 hours compared to traditional methods.
Introduction
Urinary tract infections (UTI) are among the most common infections both in outpatient and hospital settings. Over 80% of uncomplicated UTI are caused by Escherichia coli (E. coli) [1]. It is estimated that 40 to 50% of women will develop symptomatic UTI during their lifetime, and 33% will experience recurrent UTI [2]. Urine sediment analysis allows us to quickly confirm or rule out the presence of UTI. However, bacteriological analysis via culture can take up to 72 hours and is crucial for optimal antimicrobial therapy [3]. By using advanced technologies like MALDI-TOF (Matrix-Assisted Laser Desorption Ionization-Time of Flight) mass spectrometry and automated susceptibility testing from pre-treated urine sediment, it’s possible to identify pathogens and their susceptibility profiles within 24 hours. This rapid diagnosis allows for the early administration of targeted antibiotic treatment and thus facilitates patient recovery. This study evaluates the performance of direct identification (dID) and antimicrobial susceptibility testing (AST) of microorganisms from urine samples using MALDI-TOF and BD Phoenix™ M50 systems, comparing them against conventional culture methods.
Methods
The samples were obtained from patients admitted to Dr. Leonardo Guzmán Regional Hospital of Antofagasta, a high-complexity public center located in northern Chile. All urine samples included in the study corresponded to clean-catch midstream specimens. Samples were obtained from both outpatients and hospitalized individuals. The sample size was determined by the number of available samples during the study period and the limited resources provided by the grant. The inclusion of all viable samples was prioritized, although a more balanced distribution of species, such as a higher number of Klebsiella pneumoniae isolates, could have strengthened the comparative analysis. However, the representativeness of the analyzed isolates still allows for a reliable evaluation of the method’s performance for the most prevalent species in urinary tract infections. No exclusions were made based on discrepancies between dID and conventional culture identifications (cID), allowing for a comprehensive evaluation of the method’s performance in real clinical conditions. Additionally, microbial identification and antimicrobial susceptibility testing were conducted independently using MALDI-TOF and BD Phoenix™ M50, without prior knowledge of the culture results, to reduce potential measurement bias and ensure an objective analysis of the direct method. The two methodologies were performed in parallel by different personnel. Flow cytometry analysis was conducted using the Sysmex UF-5000 (Sysmex, Kobe, Japan) to determine bacterial counts. Inclusion criteria required samples to have bacterial counts exceeding 5.000/μl. This threshold was defined according to the protocol described by Zboromyrska et al. [4], classified as Gram-positive or Gram-negative based on cytometry characteristics. Samples with bacterial counts below this threshold or those containing mixed bacterial populations were excluded from the study. All samples also underwent conventional bacteriological culture for microbiological diagnosis of UTI. Samples were cultured on chromogenic orientation agar (Valtek diagnostics, Chile) and incubated at 37°C for 24 hours. Microorganism identification (ID) was performed using the MALDI Biotyper Sirius System. E. coli was identified based on chromogenic characteristics. Antimicrobial susceptibility testing (AST) was conducted using UNMIC 407 and PMIC 89 cards for Gram-negative and Gram-positive bacteria, respectively, in the BD Phoenix™ M50 system.
Urine samples were subjected to a differential centrifugation protocol to concentrate microorganisms. Initial centrifugation at 2.000 g for 1 minute sedimented larger cells and debris. The supernatant was further centrifuged at 9.000 g for 15 minutes to concentrate bacteria. The resulting sediment was washed with distilled water, recentrifuged, and used for both dID and AST, based on the protocol described by Ferreira et al. [5], with adjustments according to laboratory conditions.
The bacterial concentrate was suspended and 1 μl was deposited onto a polished steel plate for MALDI-TOF analysis following the manufacturer’s instructions. Each sample was processed duplicate using the MALDI Biotyper Sirius system (Bruker, Massachusetts, USA), and the highest score or the well that achieved identification was considered. Identification scores were classified as follows: ≥2 for species-level identification (high confidence), 1.7 to 1.9 for genus-level identification (moderate confidence), and <1.7 as doubtful. If results yielded scores ≤1.7, identification was considered valid if the top three listed microorganisms were of the same species. Control strains, including E. coli ATCC 25922, were used for quality control and instrument calibration. AST included only samples with a single microorganism identified with high confidence. Samples without valid identification were excluded, and missing values were not imputed. Non-Staphylococcus and non-Enterococcus species were excluded. Control cultures were performed to verify the purity of the microorganisms subjected to AST. Samples in which mixed growth or contamination was detected were excluded from the AST analysis to ensure the accuracy and reliability of the results. The bacterial suspension was adjusted to 0.5 McFarland and inoculated into BD Phoenix™ M50 panels according to the manufacturer’s instructions. For samples with lower bacterial densities were processed with a reduced inoculum (0.20 to 0.30 McFarland), as described by Donay et. al 6 [6].
Identification results from the MALDI-TOF system were compared with conventional culture results to assess concordance. Samples without culture results were excluded, and missing values were not imputed. Additionally, urine cultures showing polymicrobial growth by 24-hour chromogenic agar culture or direct identification of non-uropathogens were excluded from the main analysis. Cases involving mixed cultures and samples with direct identification of two microorganisms were analyzed separately. Subgroup analyses were conducted to compare the performance of direct identification and antimicrobial susceptibility testing between Gram-negative and Gram-positive. Samples were classified as follows: true positives (TP) when the same microorganism was identified by both dID and cID methods at the species level; false positives (FP) when dID identified a microorganism, but the culture was negative or cID identified a different microorganism at the species level; false negatives (FN) when cID identified a microorganism not detected by dID; and true negatives (TN) when no pathogen was detected by either method. Cohen’s Kappa coefficient with 95% confidence intervals was calculated to assess the level of agreement between direct and conventional identification methods. For a refined subset of data, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were also calculated to further evaluate diagnostic accuracy. Additionally, the performance of antimicrobial susceptibility testing was evaluated separately for each antibiotic to determine variations in agreement rates.
Susceptibility testing performance was assessed based on Cumitech 31A standards, as these were the only species with an acceptable number of samples, following the standards established by the American Society for Microbiology (ASM) in Cumitech 31A [7]. This standard establishes acceptable performance indicators when there is essential agreement (EA) and categorical agreement (CA) ≥90%, while very major error (VME) rates should be ≤3%, and major error (ME) rates should be ≤3%. For major errors (ME) and minor errors (mE) combined, the error rate should be ≤7% for at least 100 strains. To assess the reproducibility of the direct method, two representative strains—E. coli and K. pneumoniae—were processed in triplicate. Identification and susceptibility results were compared across replicates to evaluate consistency.
The project was reviewed and approved by the Scientific Ethics Committee of the Antofagasta Health Service (protocol number 0050/2024). The committee determined that, although ethical approval was required for the use of clinical samples, chart review was not necessary, as the study involved verification of a laboratory technique without direct interaction with participants and posed no ethical or legal risks
Results
A total of 473 samples were selected based on the criterion defined by flow cytometry (see Figure 1), of which 31 samples with polymicrobial culture, 13 samples without routine culture, 29 samples with direct identification of non-uropathogens, 4 samples with direct identification of two different microorganisms, and 16 samples with cultures containing 2 uropathogens were excluded. After purification, 380 samples were included in the concordance analysis: 327 from female patients and 53 from male patients. Characteristics are described in Table 1.
Flow diagram illustrating the selection and exclusion criteria for urine samples included in the study.
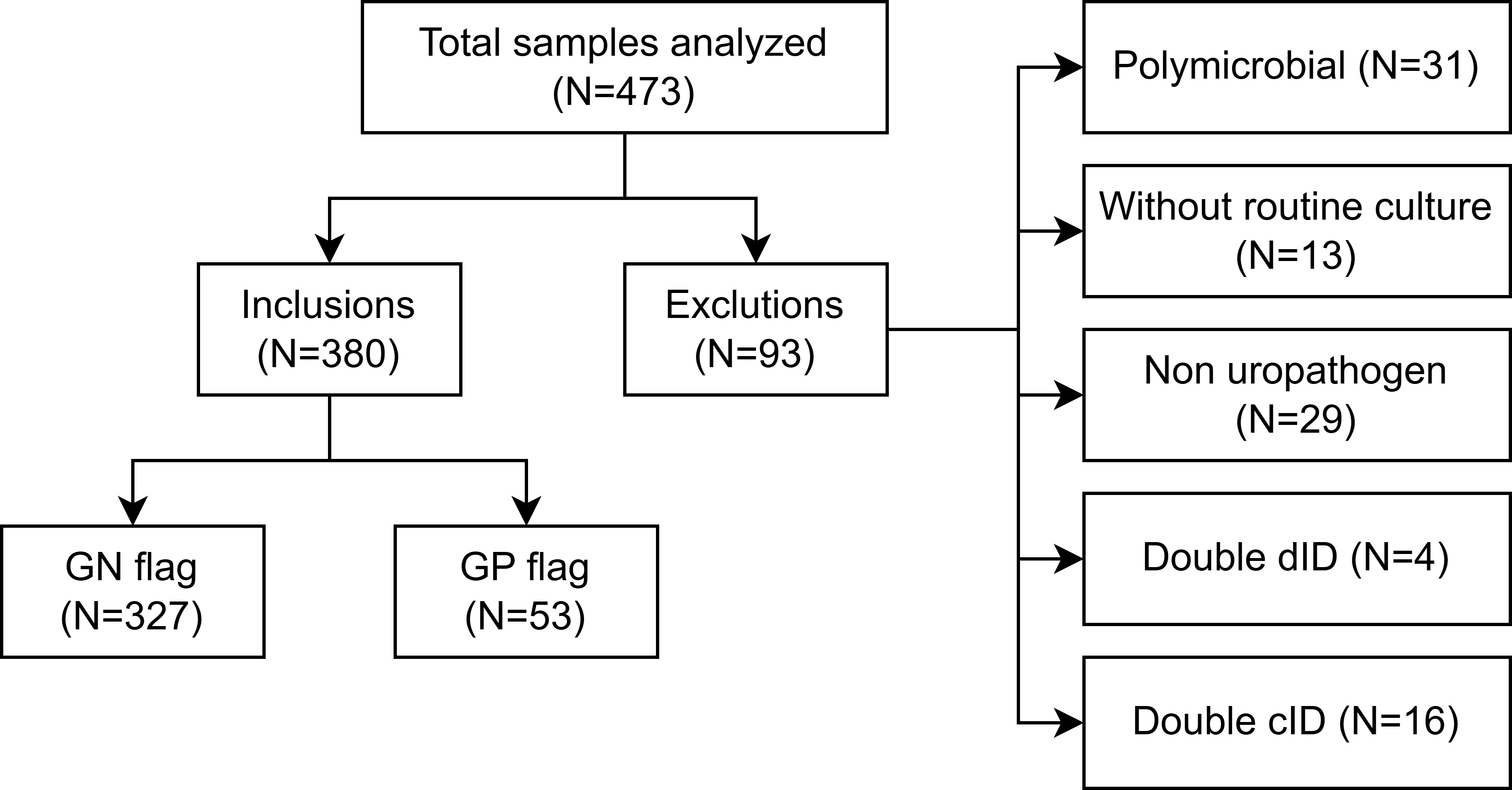
Source: Prepared by the authors of this study,
The contingency table (see Table 2) was constructed to compare performance indicators globally and by group, for both Gram-negative and Gram-positive.
The most frequently detected microorganism was E. coli (261/303) (see Table 3), with a concordance with culture of 98.46%. In four cases where E. coli was initially detected, cultures were negative; these samples had low bacterial counts (5.454 to 9.182 bacteria/µL). The second most frequent microorganism was K. pneumoniae (28/303), with a concordance rate of 96.42%. Only one discrepancy at the genus and species level was observed, in which the culture reported growth of E. coli. In both cases, identification scores were >2.0. The species C. freundii, P. aeruginosa, P. mirabilis, C. koseri, and K. variicola showed 100% concordance, although their low frequency (1 to 3 isolates per species).
In 3 cases where two microorganisms were detected via dID, complete concordance with culture was observed in only one. In the other two, dID failed to identify one of the microorganisms present. In 13 cases, the culture reported two microorganisms, while dID identify only one. In all of these, the microorganism detected by dID matches one of those identified in the culture.
In contrast, species identified by dID in the Gram-positive group are in Table 4. Concordance with culture was 100%, except for E. coli, where E. faecalis was identified in culture. Six false positives by dID were recorded. In five of these cases, cultures were negative. In the sixth, E. faecalis was identified instead of E. coli.
There were three cases in which two microorganisms were detected in culture; however, in two of them, no direct identification was achieved, and in one, dID detected E. coli, which was also identified in the culture along with E. faecalis. In only one sample were two microorganisms detected by dID (E. faecalis and E. coli), but only E. faecalis was recovered in culture.
The analysis of the results for K. pneumoniae based on Table 6 reveals variability in concordance and errors in relation to the criteria. CA and EA show that most antibiotics meet the acceptance criteria of ≥90%. However, there are notable exceptions: cefazolin, cefepime, cefoxitin., meropenem, nitrofurantoin and piperacillin/tazobactam.
In this study, due to the small number of isolates of other Gram-negative bacilli, no evaluation was conducted. For the Gram-positive analyses, there was insufficient evidence to draw conclusions, as only 11 out of 13 samples with E. faecalis identification could be analyzed, along [1] E. faecium, and [1] S. saprophyticus.
To evaluate the reproducibility of the direct method, triplicate processing of two representative strains—E. coli and Klebsiella pneumoniae—was performed. MALDI-TOF analyses consistently identified both species across replicates, achieving high-confidence scores (≥2.0) with minimal variability (mean score 2.30; range 2.28–2.35). AST demonstrated an EA of 100% and a CA of 99.2% across replicates. Minor variations of one dilution step in MIC values for cefoxitin and imipenem led to categorical discrepancies. These results indicate high intra-assay reproducibility for both direct microbial identification and susceptibility profiling under the evaluated conditions. No major errors (ME), very major errors (VME), or minor errors (mE) were observed across the triplicate AST evaluations.
Discussion
The results demonstrate an overall favorable performance of the dID method applied to urine samples. This method detects most true positive cases, making it a useful tool for initial screening of UTI, especially in the Gram-negative bacteria group, where direct method demonstrated high sensitivity and high PPV. These findings are consistent with previous studies evaluating urine and blood cultures using MALDI-TOF MS. Ferreira et al. [5] demonstrated that direct identification from urine concentrates achieves high concordance with conventional culture methods, particularly for Gram-negative bacilli. E. coli was the predominant microorganism, showing 98.46% concordance with the culture. Similarly, Klebsiella pneumoniae reaching 96% concordance. When identifications were analyzed by bacterial group, a particularly robust performance was observed in Gram-negative bacilli. These findings are consistent with previous studies by Loonen et al. [8] and Clerc O et al. [9], which report better performance of MALDI-TOF direct identification for Gram-negative bacteria from blood cultures, attributed to their more permeable cell walls and greater protein release after initial sample treatment. The specificity indicates that a considerable proportion of true negatives was also correctly identified. However, discordance was observed in 12 cases. Most of these corresponded to negative cultures, where the bacterial counts measured by flow cytometry did not exceed 12.000 bacteria/µL for both groups. This suggests that these samples may fall below the detection threshold of conventional culture, or outside the analytical criterion, which only considers counts above 1×10⁴ in midstream urine samples with abnormal sediment. Additionally, in 3 out of 11 discordant cases, discrepancies were identified at the genus or species level. This indicates that although the technique is effective in detecting bacteria, it may have limitations in achieving precise taxonomic resolution. This does not compromise its overall diagnostic value but emphasizes the need for confirmatory testing to establish the etiologic agent in clinically relevant cases. However, the NPV was moderate, suggesting that negative results should be interpreted cautiously and confirmed. The global Cohen’s kappa index indicating substantial agreement. It is important to consider that this was a conditioned sample, as most included cases were positive due to prior selection based on bacteria counts. Thus, the observed imbalance is not a design bias but an expected characteristic of the study. In this context, the kappa value may appear low—not due to poor agreement between methods, but because of the high expected agreement by chance caused by the intentional imbalance. In contrast, the high observed agreement confirms that the methods agreed in most cases.
Flow cytometry bacterial counts in the Gram-negative group showed no significant differences between TP and FN, with both groups ranging from 5000 to 999 999.9 bacteria/µL. This suggests that bacterial count alone is not a predictor of successful direct identification, and that other sample components—unassessed in this study—may interfere with the process. Bacterial count also did not correlate with higher identification scores.
The Gram-positive group showed lower sensitivity, indicating limitations in detecting true positives, but higher specificity, making it more reliable for identifying negative cultures or non-uropathogenic bacteria. This is supported by an acceptable NPV. The kappa coefficient indicates moderate agreement, while the Po, along with a much lower Pe, suggests that the method contributes diagnostic value beyond chance, and is less affected by sample imbalance compared to the Gram-negative group. This difference can be attributed to the exclusion of many samples excluded in which the direct identification revealed non-uropathogenic organisms. Among the 67 samples analyzed, only 19 (35.9%) were successfully identified by MALDI-TOF. This low identification rate is mainly attributed to technical challenges related to the cell wall structure, which impairs the generation of suitable spectral profiles without additional processing—as previously noted by Clark et al. [10]. However, 30 samples were not identified directly. Nonetheless, true positives also ranged from 5000 to 999 999.9 bacteria/µL.
Importantly, in the Gram-positive group, there were false positives in which the direct method detected Gram-negative organisms, despite flow cytometry indicating the presence of Gram-positive bacteria. We consider that these detections should be disregarded and verified by culture. In contrast, false positives in the Gram-negative group (n=6) did not show misclassification by Gram stain, but rather taxonomic errors or negative cultures—likely explained by low bacterial counts (<10 000 bacteria/µL) measured by flow cytometry.
A limitation of this study is the uncertain clinical utility of direct MALDI-TOF MS identification in suspected polymicrobial urinary infections. Although rapid, the method is less effective with multiple uropathogens, as it targets isolated organisms. In such cases, 24-hour chromogenic agar culture can complement by confirming or detecting additional pathogens.
The analysis of the results obtained in determining AST reveals both advantages and significant limitations regarding the accuracy of the method in relation to the criteria established by Cumitech 31A. One of the main benefits of these systems is the speed at which susceptibility results can be obtained, allowing for earlier clinical intervention compared to traditional culture methods. However, the reliability of these methods in all contexts is questionable, as several key antibiotics do not meet the strict criteria, requiring more evidence to ensure the reliability of these results. In the case of E. coli, a total of 13 were accepted under the criteria. These results underscore the precision of the method for certain antibiotics, while also highlighting its limitations for others, which may impact the clinical management of E. coli infections. K. pneumoniae, despite not having a significant number of samples, 8 of the 19 antibiotics in the panel met the acceptance criteria. More errors were observed in K. pneumoniae than in E. coli, with some antibiotics exceeding the maximum threshold for VME (false susceptible). The possibility of obtaining falsely susceptible results emphasizes the need for continuous validation of these systems in various clinical contexts. Falsely susceptible results can lead to inappropriate antimicrobial therapy, which can negatively impact patient outcomes [11]. It is noteworthy that certain antibiotics accepted for E. coli were not accepted for K. pneumoniae; these findings suggest that the effectiveness this method may vary between different species. It is important to note that the low number of samples evaluated limits the robustness of these conclusions. Additional studies with a larger sample size are required. The evidence obtained for Gram-positive bacteria suggests that there is a need to refine or redefine the criteria for applying this methodology in biological samples. Furthermore, direct identification using MALDI-TOF, combined with susceptibility assessment by BD Phoenix™ M50, offers the advantage of significantly accelerating diagnostic time compared to traditional culture methods. Additionally, the reproducibility analysis conducted in this study provides a preliminary indication of the method’s stability. Despite the need for further validation, the implementation of this direct method could significantly improve patient management by enabling earlier targeted antimicrobial therapy and optimizing the use of healthcare resources.

