Notas metodológicas
← vista completaPublicado el 7 de enero de 2022 | http://doi.org/10.5867/medwave.2022.01.002512
Conceptos clave para la búsqueda de evidencia: una introducción para profesionales de la salud
Key concepts for searching evidence: an introduction for healthcare professionals
Abstract
The currently abundant bibliography on healthcare can make the search process an exhausting and frustrating experience. For this reason, it is essential to learn the basic concepts of research question formulation, information sources, and search strategies to make this process more efficient and user-friendly. The search strategy is an iterative process that allows the incorporation of tools and terms in the strategy design to optimize evidence retrieval. Each strategy varies according to the questions, the language used, the source of information accessed, and the available tools. This article is part of a methodological series of narrative reviews on biostatistics and clinical epidemiology. This narrative review describes the essential elements for developing a literature search strategy and identifying the relevant evidence concerning a clinical question through familiar and accessible sources (such as Google and Google Scholar), as well as search interfaces and technical-scientific databases focused on biomedical knowledge (PubMed and The Cochrane Library).
Main messages
- Formulating a well-defined clinical question is crucial to perform an effective literature search
- When formulating a question, colloquial or everyday language ("natural language") and other languages specific to databases ("controlled language") can be used.
- Search strategies are iterative and optimizable processes. According to the results obtained, multiple searches and modifications should follow one after the other to refine the quality and quantity of results
- Iterations enable a balance between sensitive and specific searches through various basic and advanced tools (field labels, filters, truncations, and wildcards, among others).
Introduction
Evidence-based medicine (EBM) is a tool that seeks to converge the best available evidence with clinical expertise, in addition to values and patient preferences [1]. Traditionally, five steps are described for a practical implementation, known as the five As. These are: ask (formulate the question), acquire (search for the best evidence), appraise (evaluate findings), apply (apply the results), and assess (evaluate the results) [2].
Nowadays, the available information on a given topic can become very broad, resulting in an overwhelming and burdensome process if the necessary tools are unavailable or unknown. It is estimated that only half of healthcare professional questions are searched, and only about two-thirds of these are answered [3]. The main challenges encountered by physicians are the lack of training in bibliographic search, the lack of knowledge of the multiple digital sources of information, and poor search strategies [3].
This article is part of a methodological series of narrative reviews on general biostatistics and clinical epidemiology topics, which explore and summarize published articles available in the main databases and specialized reference texts in a user friendly language. The series is oriented to the training of undergraduate and graduate students. It is carried out by the Chair of Evidence-Based Medicine of the School of Medicine of the 'Universidad de Valparaiso', Chile, in collaboration with the Research Department of the University Institute of the 'Hospital Italiano de Buenos Aires', Argentina, and the Catholic University Evidence Center. This narrative review describes the essential elements for developing a literature search strategy and identifying relevant evidence concerning a clinical question.
The objective of the literature search
The main objective of a literature search is to find scientific articles and other sources of information that can answer a focused clinical question while making a practical, structured, and reproducible search.
The search should begin once the critical elements of the clinical question have been defined. If the clinical question is not formulated correctly, the search will probably not yield good results. Consequently, it is essential to formulate a well-defined question that allows us to establish criteria for selecting the evidence and approach the results of our interest. For practical purposes, we can classify the questions into:
a) Fundamental or global deficit (background) questions are general questions on basic academic concepts about a given disease or clinical concept [4]. For example: What is the pathophysiology of heart failure? What are the possible differential diagnoses for a given symptomatology? The ideal sources of information to answer this type of question can be textbooks, narrative reviews, or bibliographic abstracts. These are the so-called secondary sources of information [4].
b) Advanced or foreground questions are questions concerning specific knowledge applied to a particular patient or problem [4]. The PICO format is the most widely used to answer foreground questions. The letter "P" refers to the population or patient we are interested in and its characteristics. The "I" corresponds to the intervention we seek to carry out, the "C" is the comparison (therapeutic alternative or placebo), and the "O" is the objective or outcome we wish to evaluate [4]. For example: "In infants with bronchiolitis (P), does treatment with salbutamol (I) compared with placebo (C) decrease overall mortality (O)?". This type of question focuses on treatment, but there are also modifications to the PICO approach depending on whether the question is prognostic, etiologic, diagnostic, prevalence, or harm-focused (adverse events). For didactic purposes, we take therapeutic questions as a model in this article.
Sources of information
Knowing and understanding the available resources to answer a specific clinical question allows us to search more effectively and efficiently. This search is intended to lead to the most appropriate answer for the clinical context in which it is developed. A clear example can be seen in the abovementioned questions. Background questions can be answered by searching secondary information sources (textbooks, encyclopedias, or bibliographic summaries). On the other hand, foreground questions require an exhaustive search from many sources (scientific journals and medical bibliography databases). In the latter, the amount of information can be substantial, and the quality can be varied. Considering this aspect, Haynes proposed the "6S pyramid", a model in which he organized the different types of evidence (Table 1) [5],[6].
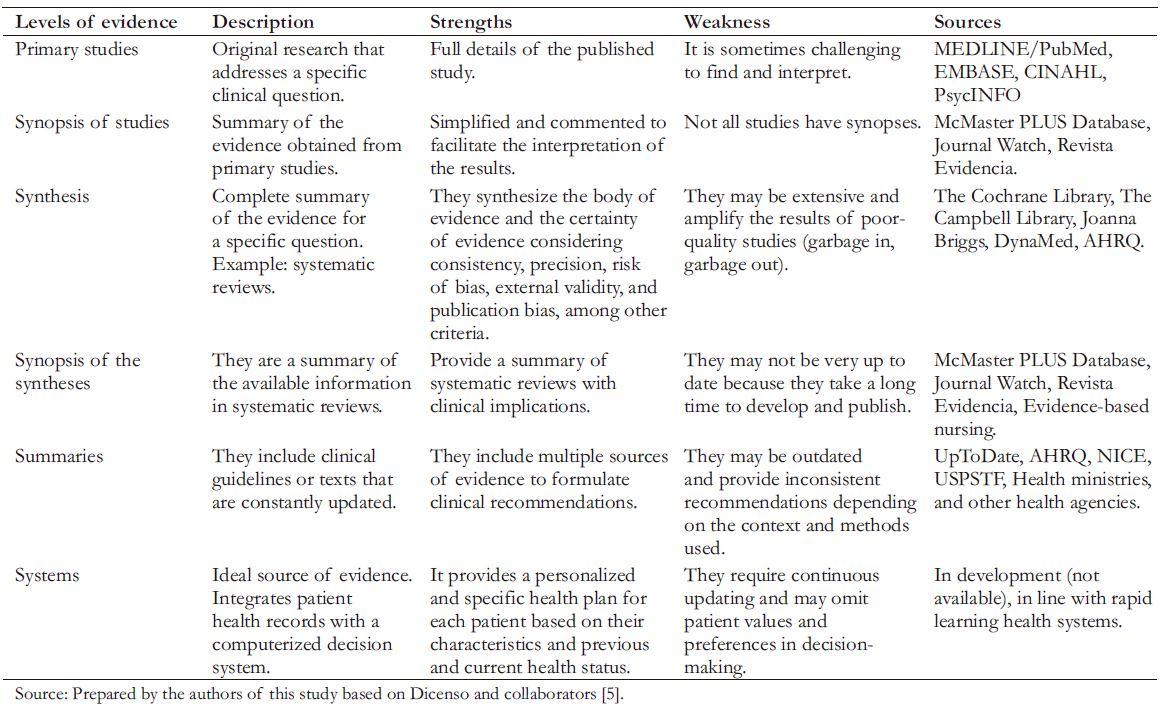 Full size
Full size Once a clinical question has been formulated, and the vital source of information has been identified, we can move on to the next step. At this stage, we need to select the necessary databases, evaluate the availability of search interfaces, design the search strategy and adapt it to the interface(s) that allow us to retrieve the information relevant to our question.
In the following sections, we describe the methodological process of a bibliographic search, considering the types and characteristics of the information sources. Specifically, we address the search tools that are most used in Latin America. In Box 1, we present a glossary of the terms used in the article. Some specialized resources (EMBASE, PsycINFO, and CINAHL) will not be addressed because they are relevant to specific areas of knowledge, require institutional subscriptions, and are not the most frequently used by health professionals.
Database A set of structured and classified information set whose purpose is to preserve and retrieve information. Reference databases provide bibliographic information (e.g., authors, title, year) of the indexed journals. Examples: MEDLINE, LILACS, Scopus, EMBASE, CINAHL, PsycINFO, CENTRAL.
Thesaurus/controlled language A list of words or terms used to represent concepts used in a given database. Examples: MeSH for MEDLINE and DeCS for LILACS.
Natural language Words that are used daily in clinical practice. It includes those terms that are not predefined in the database or authors use in their publications. In some manuals, they are called "text terms".
Boolean operator Words that connect search terms by establishing relationships between them. Boolean logic allows to create, combine, or discard terms according to the three criteria: "AND", "OR", and "NOT". Combining search terms with Boolean logic enables the user to design and execute search strategies in a database. The possibilities of designing a search strategy depend on the database and its interface because they have predefined tools that allow different search modes. However, some basic Boolean operators ("AND", "OR", "NOT") are universal and allow to join the search terms of the query logically.
Search interface An entry point that allows access to the contents of various information resources. By combining keywords, thesaurus terms, and Boolean operators, the interface allows access information from databases. For example, the MEDLINE database can be accessed through the Ovid or PubMed interface. In turn, Google is an interface that can search information from multiple databases, including MEDLINE.
Indexing The process of identifying a thematic description of a document with terms representing its content in a database.
Repository A digital archive where digital information is collected, stored, organized, and disseminated as scientific and academic material. Examples: university academic repositories, SciELO, and PubMed Central (PMC).
Gray literature Scientific-technical information not formally published in sources such as books or journal articles. Example: clinical trial registries (clinicaltrials.gov), government documents (including health technology assessments and clinical practice guidelines), and academic theses. This documentation can be found in institutional repositories.
Source: Prepared by the authors of this study.
How to search in google/ google Scholar
Google’s search interface is the most widely used because it is intuitive and accessible. Google uses artificial intelligence to
search, index, and retrieve the most relevant results [7]. We likely find synthesized and simplified information intended for patients or the general population using natural or colloquial language. Instead, technical language, preferably in English, should be used to find scientific information (e.g., types of research design such as clinical trials or case-control studies). Some strategies to obtain better results include:
a) The use of scientific language.
b) Describe the methods sought (e.g., study design).
c) Add terms related to reliable databases/interfaces (PubMed, SciELO, Cochrane, or others).
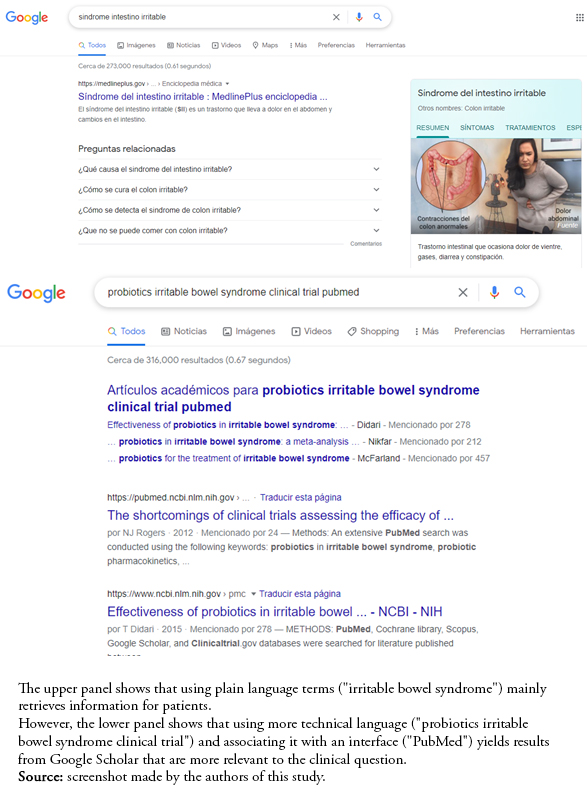 Full size
Full size Suggestions usually appear from the Google Scholar interface (https://scholar.google.com/). The algorithms used by the conventional Google search engine are different from those of Google Scholar since the latter prioritizes searching and indexing complete and original articles based on relevance, with less advertising and commercial priority. It has the advantage of having a wide variety of bibliographic material, including books and articles.
Google Scholar uses more frequently non-English literature (up to 40%) for its citation count, unlike other databases (Web of Science and Scopus), whose frequency of English literature reaches up to 90% [8]. In addition, it indicates how many times and by whom an article has been cited. It is a source that allows access to information resources from different areas of knowledge, retrieving results not found in area-specific sources [9]. It has limited search support from Boolean operators and other search operators as a disadvantage. In addition, the ranking of search results considers the availability of full text, place and site of publication, author(s), and citation frequency. However, the relative weight of these factors is unclear [10].
Finally, the search options are inflexible (it is not possible to filter by document type, search by field, or refine by subject, among others), and its instructions are not very accessible. There is also no rigorous quality control of the sources processed, and duplicates may be found. For these reasons, it may be necessary to resort to the advanced search option, which filters the results by date, author, and article content. Another option this platform provides is linking the search to university libraries [11] (Figure 2).
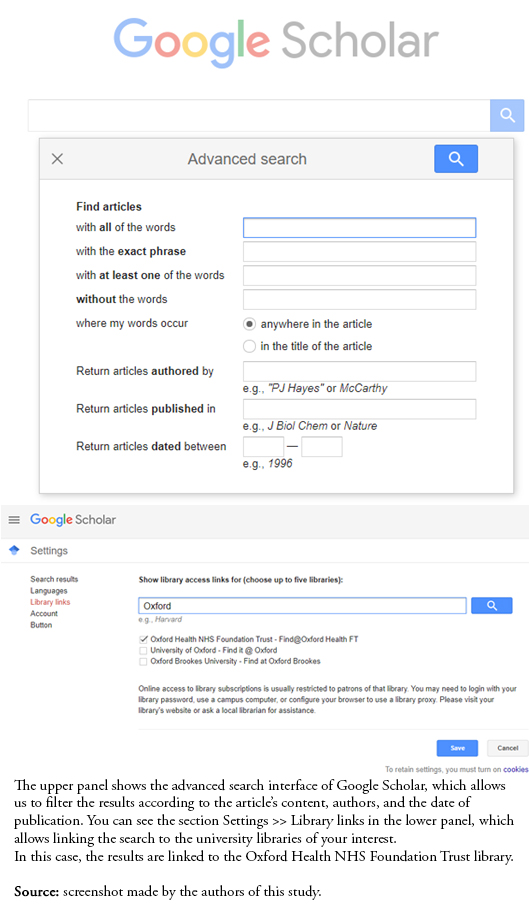 Full size
Full size How to search in PubMed (Medline)
PubMed (https://pubmed.ncbi.nlm.nih.gov) is the search interface of the US National Library of Medicine that allows free access to more than 30 million bibliographic citations, including the MEDLINE database – one of the most important databases in health sciences [12]. The results are more
appropriate for a healthcare-related search than a basic Google search as this engine focuses on biomedical knowledge. Its tools allow advanced, reproducible, and more controlled searches, meaning the retrieved results will be similar using the same search strategy. However, the interface is not intuitive for people unfamiliar with Boolean operators. PubMed retrieves
bibliographic data to access the full text, including the digital object identifier (DOI). The DOI allows searching the document in different information resources to access its contents and add the bibliographic data in managers such as Paperpile and Zotero [13].
PubMed has two types of searches: basic and advanced. Enter the term or phrase of interest and press the Search button to start a basic search. PubMed uses the "AND" operator between word spaces in a search phrase by default. Therefore, we must check that there are no extra spaces between terms and correct logical operators to perform the search.
PubMed: basic search
When searching in PubMed, we must consider the language used. Natural language is preferred if we want a more sensitive search. If we want the search to be more specific, we must use the controlled language of the MeSH thesaurus. Another feature of the controlled language is that it includes all the synonyms and variants of the search term, considerably reducing the risk of losing articles that do not include the specific term used in the search strategy. For example, "Renal Insufficiency" [MeSH] retrieves articles that cover "renal insufficiency", "kidney insufficiency", "renal failure", "kidney failure", among others. It is important to note that these languages (natural and controlled) are complementary, and it is advisable to use both in the same search (Table 2).
 Full size
Full size PubMed: advanced search
The advanced search is helpful for a structured and reproducible approach. In these cases, natural and controlled language terms are selected a priori, corresponding to each concept that composes our query. By identifying the search interface and its possibilities, the terms can be added and combined in specific bibliographic records fields, combining them with the Boolean operators that best suit our search. In addition, there are additional tools such as filters, wildcards, and truncations, which allow refining the search even further (Box 2).
Field labels Each field of a bibliographic record (author, title, abstract, others) is identified by a label of two or more letters that can be added after each term in square brackets ([AU];[TI];[AB]; respectively). This tool allows performing a search exclusively in that field. For example: When searching for "Heart failure[TI/AB]", we retrieve all records with the term "Heart failure" within the title and abstract fields.
Filters These are tools available in the search interfaces to increase precision. For example: PubMed allows to narrow a search using filters, located on the left side of the interface, according to publication date, article type, and text availability, among many others.
Truncation (*) These are used to replace a character or a set of characters to the right of a term. They are used to search for term derivatives (suffixes and grammatical inflection). The asterisk (*) is used as a truncation character on the right to find all word forms. For example: child* retrieves all documents that include children and variables that follow the letter "d", e.g., child, children, childhood. The Ovid platform allows truncating and retrieving variants with the symbols "$" or ":". For example, we can use either child$ or child:, as these symbols fulfill the same function.
Wildcardsa Other characters such as the numeral (#) allow us to replace one character inside or at the end of the word. For example: wom#n. ti retrieves references in the title whose word is woman or women. The wildcard represented by the question mark (?) performs a similar function to the previous one, with the difference that it replaces one or no character. This tool helps retrieve terms spelled differently in American and British English. For example: h?ematology retrieves all papers including hematology and haematology.
NEAR or NEXTa proximity operators The former specifies that the included terms should appear close to each other in any order. For example: if we search for "cancer NEAR lung", the retrieved results may include "lung cancer" AND "cancer of the lung" (separated by up to six words between the terms). If we want to reduce the number of terms, we can use the NEAR/x command, where x is the maximum number of words between search terms. Furthermore, the NEXT proximity operator states that the terms must appear in order and assumes terms close to each other. If we apply the same example: "lung NEXT cancer", the results retrieved would be "lung cancer" or "lung metastatic cancer" but not "cancer of the lung". (a)
These functions are not available in the PubMed platform.
Source: Prepared by the authors of this study.
Targeted or sensitive search
These multiple tools available in PubMed allow a more flexible search. If we want an even more specific search (i.e., as few results as possible and more relevant to our query), we should add more components of the predefined PICO query linked with the Boolean operator "AND". It is preferred not to use the Boolean operator "NOT" because relevant references may be excluded. In addition, filters available in the interface (e.g., for systematic reviews or clinical trials) can be used to narrow the search and further decrease results based on the study design (T) component of the PICO question [14]. Some
generally inadequate methods used to increase search specificity include:
a) Limiting by date: relevant articles may be published before the imposed limit. However, it would be reasonable to limit by date to identify systematic reviews since older ones may be outdated.
b) Limit by article availability ("free full text" in PubMed): the relevance of an article is not determined by its accessibility. In these cases, it is suggested to consult with your institution to obtain the full text of the reference.
Suppose we want a more sensitive search (with the largest possible number of results, but with many results that may not be relevant to our question). In that case, we can remove components of the PICO question and apply the least number of filters (or omit them). Other options to broaden the search are the application of truncations and adding additional terms with the Boolean operator “OR”.
It is essential to understand that search is an iterative practice: the process does not end with a single search. In contrast, we need multiple searches to refine the quantity and quality of results obtained from analysis and the aforementioned tools. Ultimately, the sequence of searches, the evaluation of the results, and the appropriate use of the available tools determine the final accuracy.
Steps of an advanced search design
Once the advanced search structure has been created, the first step is conceptualizing the PICO question’s elements with controlled language (MeSH) and natural language. We need to use natural language terms in the title and abstract fields ([title/ abstract] or [tiab]) to avoid irrelevant results. Terms that relate to the same element of the PICO question are joined by an "OR" and closed in parentheses, thus forming the search line (e.g., line number 1 or #1). We should repeat this process with all the elements until the lines join the final strategy with an "AND". Finally, the results obtained by our search are displayed and analyzed. In case the retrieved results are not as expected, it is essential to review the literature on the topic we are trying to address and apply the tools described in the previous section to refine the sensitivity and specificity of the search (Table 3).
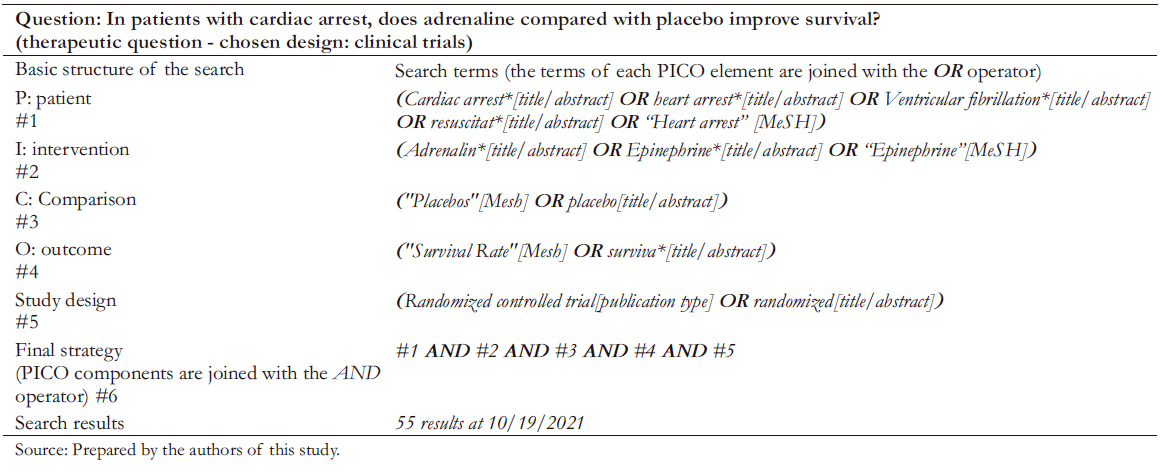 Full size
Full size How to search in the Cochrane library
The Cochrane Library (https://www.cochranelibrary.com) is a collection of databases containing high-quality evidence for decision-making in healthcare. This platform has more than 8300 reviews, 2400 protocols, 1 600 000 clinical trials in the Cochrane Central Register of Controlled Trials (CENTRAL), 2400 Cochrane Clinical Answers (brief summaries of Cochrane systematic reviews), 300 000 Epistemonikos reviews, 130 editorials, and 30 special collections. In addition, it has the advantage of being available in multiple languages, including English and Spanish [15].
Like PubMed, it has two search options: basic and advanced. Type the term of interest in the search box and select the
desired search field to start a basic search. The Cochrane Library also allows searching by topic or Cochrane Review Group. The logic is similar to an advanced search in PubMed. However, it is worth mentioning that The Cochrane Library has a search system based on the PICO format, which is currently in the testing phase and is limited to Cochrane systematic reviews. In addition, the advanced search adds other search tools, such as the proximity operators (Box 2).
In brief, the Cochrane Library has a search system similar to PubMed and a wide range of information content. Mainly, it is a source that can be accessed when our objective is to obtain a complete summary of the available evidence on our clinical question.
How to search in lilacs
LILACS (https://lilacs.bvsalud.org) is a database that includes technical-scientific papers produced by authors from Latin America and the Caribbean related to health sciences. This database has its thesaurus called DeCS, which is based on MeSH terms and is available in English, Spanish, and Portuguese [16],[17]. The methodological process is similar to the other information sources seen above, although advanced searches are limited. LILACS has an interface with tools and filters specific to the information source. Generally, we suggest using a few search terms and natural language (Table 4).
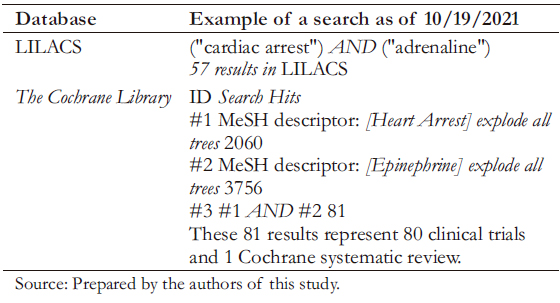 Full size
Full size Conclusion
To find the best available evidence for a focused (foreground) clinical question, we need to determine the type of question (therapeutic, prognostic, etiological, diagnostic, prevalence, or harm), identify its components, and apply them to the PICO format or its variants. Once our question has been formulated, it is necessary to determine which source of information (6S pyramid) is the most relevant for obtaining the results. And finally, after selecting search locations, we must always keep in mind that a search process is iterative. It is necessary to try different variants in the search strategy – using natural and controlled language and tools such as truncation, Boolean operators, filters, among others – until we obtain satisfactory results.

