Artículo de revisión
← vista completaPublicado el 20 de octubre de 2025 | http://doi.org/10.5867/medwave.2025.09.3026
Evidencia disponible de integración de COVID-19 a sistemas de vigilancia centinela: revisión de alcance
Available evidence on integrating COVID-19 into sentinel surveillance systems: A scoping review
Abstract
Introduction The COVID-19 pandemic exposed the weaknesses of epidemiological surveillance systems and highlighted the need to integrate new respiratory viruses into sentinel surveillance systems. However, current evidence on their effectiveness remains limited.
Aim This project conducts a scoping review to describe the available evidence on the integration of COVID-19 into sentinel surveillance systems.
Methods The included studies addressed sentinel surveillance in the context of the pandemic following the World Health Organization declaration. A systematic search was performed in databases including MEDLINE, LILACS, EPISTEMONIKOS, and DIMENSIONS, selecting observational studies and systematic reviews. Data collection and analysis were organized into categories such as clinical characteristics, timely detection, geographic representativeness, co-infection, and adaptability with genomic surveillance. Seventeen studies reporting on COVID-19 integration impact and one preliminary WHO report were identified.
Results Results identified the most prevalent symptoms in the general population: fever (73%), cough (51.8%), loss of taste or smell (45.1%), hypoxemia (33%), and sputum production (23.9%). A high correlation was obtained between SARI cases or hospitalizations due to respiratory infection and the incidence of COVID-19 (ρ = 0.78 and ρ = 0.82 respectively).
Conclusions Integrating COVID-19 into the sentinel surveillance system could improve detection, response, and follow-up capacity. Additionally, implementing standardized case definitions promotes more efficient use of laboratory resources, thereby enhancing the sustainability of the surveillance system.
Main messages
- The scoping review focused on examining the available evidence for sentinel surveillance systems, including clinical characteristics, timely detection and response, geographic representativeness, efficient resource utilization, and adaptability in the context of genomic surveillance.
- The integration of COVID-19 into sentinel surveillance systems for severe acute respiratory infections has shown that it could have a favorable impact.
- Despite the observed benefits, the available evidence is limited and heterogeneous due to differences in sentinel surveillance systems in different countries.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), first reported in Wuhan, China, in late 2019, triggered the COVID-19 pandemic, which impacted respiratory infection surveillance activities. Currently, many countries are transitioning out of a state of emergency, following the recommendations of agencies such as the World Health Organization (WHO) and the European Centre for Disease Prevention and Control (ECDC) [1]. Sentinel surveillance is based on information from "sentinel units", which report on a predetermined sample of individuals in whom the event of interest occurs [2]. The WHO recommends specific case definitions, including Acute Respiratory Infection (ARI), Severe Acute Respiratory Infection (SARI), and Influenza-like Illness (ILI) [3]. Additionally, the information is crucial in public health. The data obtained can guide strategies to limit the spread of SARS-CoV-2, as well as to reduce morbidity and mortality. WHO suggests maintaining and strengthening sentinel surveillance [4]. Finally, efforts have been initiated to assess priorities from a country perspective, identifying challenges and gaps in integrating COVID-19 into surveillance systems for influenza and other respiratory viruses [5].
In this context, the integration of COVID-19 into sentinel surveillance systems for severe acute respiratory infection represents a key tool for more efficient monitoring against future pandemic threats. This project aims to describe the available evidence on the impact of integration, analyze the responsiveness of epidemiological surveillance systems, monitor virus variants, and evaluate health interventions.
Methods
This review followed the five stages described in the Arksey and O'Malley framework [6].
Stage 1: identification of the research question
The following question directed this structured review:
What is the available evidence of the impact of COVID-19 integration into sentinel surveillance systems on COVID-19 management?
Phase 2: identification of relevant studies
A systematic search was conducted in the following databases: MEDLINE/PubMed, LILACS, Epistemonikos and Dimensions. The latter incorporates artificial intelligence, which optimizes information retrieval by applying machine learning algorithms and natural language processing. The search strategy used in Dimensions was: ("COVID-19" OR "coronavirus" OR "SARS-CoV-2") AND ("sentinel surveillance" OR "surveillance system" OR "sentinel networks" OR "sentinel surveillance system" OR "sentinel surveillance system" OR "sentinel surveillance system" OR "sentinel surveillance system") AND ("Universal Surveillance" OR "Mass Surveillance").
After selecting the studies, a literature review of the references from the selected articles was conducted. The systematic search was limited to publications in English and Spanish, and focused on studies published since 2019. In addition, a search of gray literature was included in technical and operational reports issued by public health agencies, such as the WHO, and by governmental entities. Incomplete studies, opinion articles, narrative reviews, articles that did not include the impact of COVID-19 integration into sentinel surveillance systems, and studies conducted outside the temporal context of the SARS-CoV-2 pandemic were excluded.
Phase 3: study selection
Two authors (JGS and JAI) independently reviewed titles and abstracts. They then reviewed relevant articles in their entirety. Articles describing the integration of COVID-19 into sentinel surveillance systems and case definition criteria confirmed by reverse transcriptase polymerase chain reaction (RT-PCR) were included. If the two reviewers could not agree on the inclusion of the abstract or the full article, the opinion of a third reviewer (DPM) was sought.
Phase 4: data charting process
Two reviewers (JGS and DPM) developed the standardized matrix in Microsoft Excel ® (Annex 1). Two authors (JGS and JAI) independently performed the data extraction. Information related to general data (title, year of publication, country) and methodological data (research design, objective and clinical outcomes) was collected. The critical appraisal tools of the Joanna Briggs Institute (JBI) [7] were applied to analyze the reliability, relevance, and results of the included articles.
Phase 5: summary of results
The results were organized into the following categories: clinical characteristics, timely detection and response, geographic representativeness, efficient use of resources, coinfection, and adaptability with genomic surveillance. The review was conducted according to the Preferred Reporting Items for a Systematic Review and Meta-Analysis guidelines extension for scoping reviews (PRISMA-SCR) [8].
Results
We identified 44 studies exploring the integration of COVID-19 into sentinel surveillance systems (Figure 1).
Flow diagram according to the PRISMA-ScR statement for integrating COVID-19 into sentinel surveillance systems.
Source: Prepared by the authors based on the results of the study.
The included studies were: 1 systematic review and 16 cross-sectional studies from: United Kingdom [9], Spain [10,11,12], Belgium [13], Kenya [14], China [15], United States [16,17,18], Portugal [19], Egypt [20], Israel [21,22], Uganda [23], Niger [24], Bangladesh [25] and a WHO report [26]. The studies reported data between January 30, 2020, and December 15, 2023 (Table 1).
Epidemiological surveillance of COVID-19 has represented a global challenge for health systems, requiring adaptive and efficient approaches to its monitoring. Among the systems implemented, sentinel surveillance and universal surveillance have been widely discussed in terms of their performance, sustainability and capacity to generate relevant information in diverse contexts. Based on the evidence available between 2020 and 2023, eight key characteristics have been evaluated: sustainability, geographic representativeness, coinfection monitoring, resource efficiency, early detection, variant detection, cost-effectiveness and sequencing capacity. The results are classified as positive, potentially positive, and inconclusive, with the amount of evidence supporting each finding also indicated (Figure 2).
Comparison of COVID-19 surveillance systems. Sentinel versus universal evidence-based surveillance 2020 to 2023.
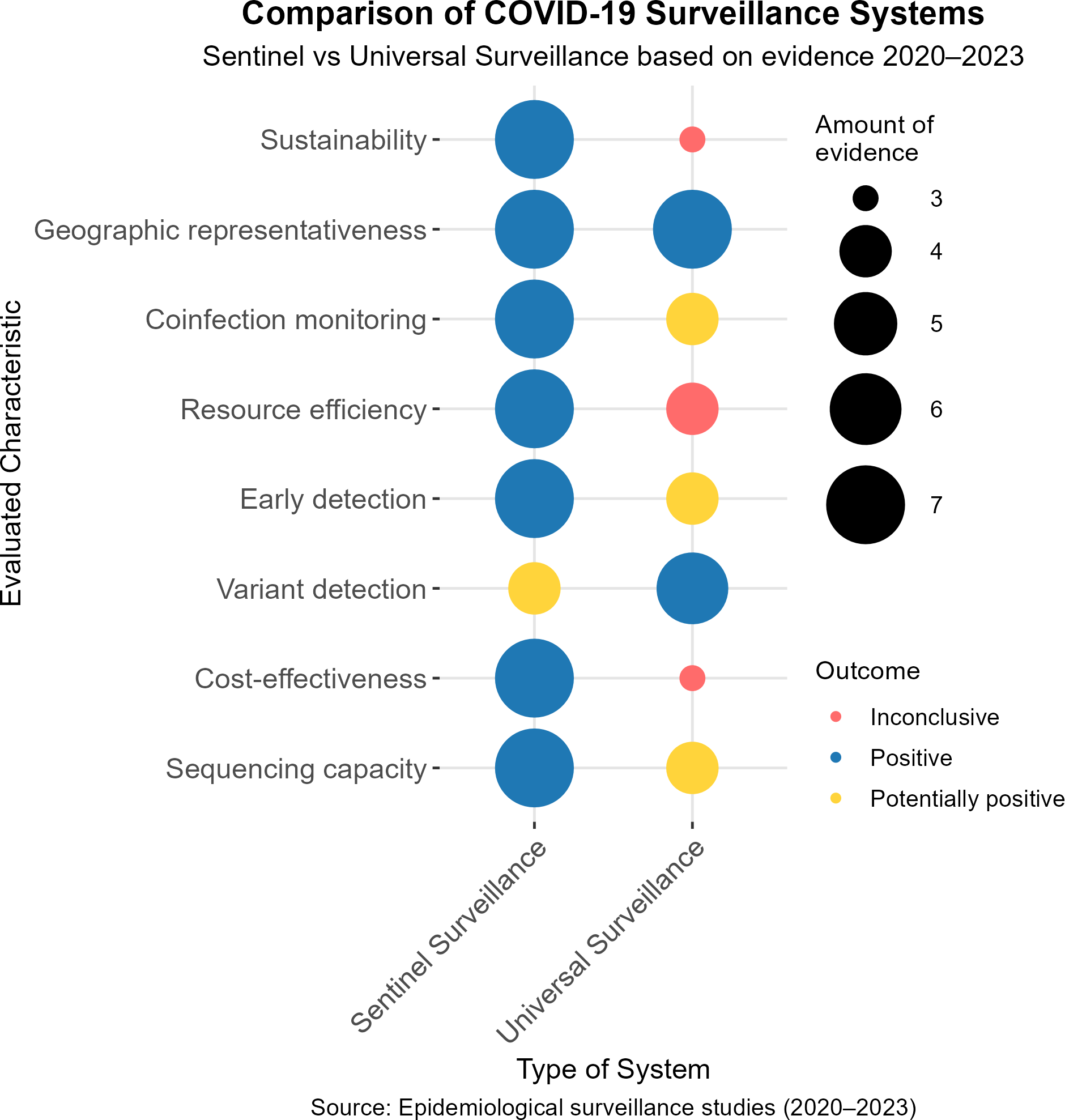
Clinical features
The frequency of signs and symptoms constitutes a crucial element in developing a case definition within the framework of sentinel surveillance. In this context, the results of three studies analyzed the clinical characteristics of COVID-19 in different geographical settings. These findings highlight fever and cough as the most prevalent symptoms, although significant differences are observed according to the characteristics of the population and the setting evaluated (Table 2).
Additionally, the WHO, in its latest publication on the strategy for integrating COVID-19, reported sensitivity and specificity based on the case definitions used for influenza surveillance, yielding similar values for comparison (Table 3).
Timely detection and response
Studies conducted between 2022 and 2023 have demonstrated that sentinel surveillance enables the early identification of epidemic peaks, concurrent circulation of respiratory pathogens, and SARS-CoV-2 positivity rates. In addition, these have provided essential information for public health decision-making and the design of epidemiological control strategies (Table 4).
Geographical representativeness
Surveillance systems require samples to be representative of the general population, as demonstrated by a study conducted in Washington, USA, which analyzed the geographic extent of available sequencing data for COVID-19 cases using a hotspot map. Prior to the implementation of sentinel surveillance, only 3.3% of confirmed cases (10 653 of 323 121 total cases) had sequencing information available, which limited the representativeness of the geographic data. However, during the period when sentinel surveillance was implemented, this percentage increased to 12.1% (56 106 cases sampled). This allowed for greater coverage and uniformity across counties. These results highlight the significant contribution of sentinel surveillance to improving the representativeness and utility of infectious disease monitoring systems.
Efficient use of resources
In three studies from Spain, it was observed that in the autonomous community of La Rioja, a comparison between sentinel and universal surveillance systems for monitoring acute respiratory infections (ARI) reveals that, although general trends show consistency, the sentinel surveillance system presents variations. For example, in children under four years of age, sentinel surveillance reached higher incidence peaks (4958 cases per 100 000 population compared to 3691 in universal surveillance), while in week 2 of 2022, it reported 3153.4 cases per 100 000 population compared to 1773 in universal surveillance. These differences in incidence reflect the ability of sentinel surveillance to capture epidemiological patterns. This enables the optimization of resources by reducing the need to monitor the entire population. In Castilla y León, 62 000 people (2.6% of the total population) were monitored by 68 strategically distributed basic surveillance units. This approach enabled the efficient capture of epidemiological trends and reduced operating costs, ensuring the sustainability of the surveillance system.
Coinfection
Coinfection with SARS-CoV-2 and other respiratory pathogens, such as influenza A, influenza B, and respiratory syncytial virus, is a clinical/epidemiological problem in East Africa, as evidenced by studies in Uganda and Kenya. During the study period, a total of 22 cases of coinfections were detected by surveillance centers for influenza/severe acute respiratory infection type diseases: 2 cases with SARS-CoV-2/AH3, 9 cases with SARS-CoV-2/B.1.1.7 (also known as B. 1.1.7 or B. 1.1.7), and 11 cases with SARS-CoV-2/AH1pdm09. On the other hand, in Kenya, out of a total of 1271 individuals, the most common co-infectious pathogens were Streptococcus pneumoniae (n = 29) and Haemophilus influenzae (n = 19), which accounted for 2.3% and 1.5% of all SARS-CoV-2-positive samples, respectively.
Adaptability with genomic surveillance
The ability to rapidly detect new pathogens is a key indicator of the effectiveness of a surveillance system. For example, a study conducted in Belgium with 5695 respiratory samples found that 1558 samples tested positive for SARS-CoV-2, and 925 of these samples were successfully sequenced.
Initial genomic surveillance enabled the evaluation of the epidemic growth rate of SARS-CoV-2 variants by examining the growth slopes during the epidemic phase. In these, BA.1 (23.21) was the highest, followed by B.1.617.2 (Delta, 13.87), BA.5 (12.10) and BA.2 (10.91), while P.1 (1.84) and BA.4 (1.92) had much slower growth. Variants such as B.1.351 and BA.3 showed no definite epidemic growth (R² indeterminate), and P.1 had a poor regression fit (R² = 0.90). Although B.1.617.2 achieved 100% weekly detection, other variants had varied peak detection levels, such as BA.2 (98.5%), BA.1 (95.3%), and B.1.1.7 (85.1%), in contrast to BA.3 (0.14%) and B.1.351 (4.6%). The period between initial detection and epidemic growth varied between 2 and 8 weeks, depending on the variant. This highlights the importance of early surveillance to identify rapidly spreading variants.
In Israel, genome sequencing, performed on sentinel samples positive for SARS-CoV-2, provided additional support for this same sentinel surveillance. In Niger, out of a total of 51 SARS-CoV-2 positive samples identified through the sentinel surveillance system, 23 (45.1%) were eligible for sequencing. This analysis revealed the identification of two Omicron sublineages, notably BA.5 and BA.3, as well as 14 XBB.1/XBB.1.5 sublineages and one recombinant XBD variant. The XBB.1.5 sublineage is of considerable concern due to its rapid spread in the United States, underscoring the importance of identifying and characterizing it through genomic surveillance. The latter demonstrates the ability to detect specific sublineages and recombinant variants, allowing monitoring of their spread and behavior.
Discussion
The review demonstrates that the integration of COVID-19 into sentinel surveillance systems has improved the ability to monitor respiratory diseases. It has also enabled the early detection of epidemic thresholds and the identification of coinfections [27,28]. In this line, it could be an efficient tool to optimize resources, especially in scenarios where health systems faced economic constraints during the COVID-19 pandemic. This was particularly relevant in some countries, where the redirection of resources allowed for improved surveillance of high-incidence respiratory diseases [29].
The implementation of multiplex PCR tests has enabled the more accurate identification of coinfections between SARS-CoV-2 and other respiratory pathogens. Similarly, they have been useful for monitoring the simultaneous circulation of different respiratory viruses [30,31].
One of the primary challenges is integrating genomic surveillance into sentinel surveillance. More than 50% of countries already perform genetic sequencing, which offers a cost-effective and sustainable opportunity to track SARS-CoV-2 variants and strengthen the response to future pandemics [32].
The economic and operational implications vary significantly between countries, depending on their health systems and level of development. In countries with advanced health systems, such as Israel and Portugal, the preexistence of technological infrastructure, specialized personnel, and integrated digital health information systems facilitated the simultaneous implementation of sentinel and genomic surveillance for COVID-19. These countries achieved high sensitivity in detecting variants and epidemiological changes, as evidenced by the significant correlations reported in Portugal between cases of severe acute respiratory infection and the incidence of COVID-19. The capacity for genomic sequencing in these settings enabled the accurate characterization of circulating viral diversity and the early detection of emerging variants. This differs from resource-limited countries such as Niger and Bangladesh, which face significant challenges, including inadequate diagnostic infrastructure, limited information systems infrastructure, and a shortage of skilled personnel. These countries had to adapt their strategies by selectively prioritizing sentinel sites, reducing geographic coverage but maintaining population representativeness. In Niger, for example, only 45.1% of SARS-CoV-2-positive samples were eligible for sequencing, which could result in an incomplete characterization of circulating viral diversity. Despite these limitations, they successfully implemented functional systems that provided valuable epidemiological information for local decision-making.
Heterogeneity in sentinel surveillance systems across countries constitutes a significant methodological limitation for this review. Differences in operational case definitions (influenza-like illness, acute respiratory infection and severe acute respiratory infection), sentinel site selection criteria, sampling protocols and diagnostic algorithms employed make direct comparability of results difficult. For example, while some countries, such as Spain, implemented systems with high geographical representativeness (68 basic surveillance units in Castilla y León), others had to concentrate resources in specific areas. Likewise, the variability in genomic sequencing capacity (from less than 50% in low-resource countries to nearly 80% in developed countries) generates asymmetries in the detection and characterization of variants. Future studies should develop standardized frameworks for evaluating surveillance systems that explicitly address these contextual differences, allowing more robust comparisons between diverse settings.
Conclusion
Finally, WHO’s interim guidance on integrating SARS-CoV-2 and influenza surveillance underscores the need for flexible surveillance systems, with strong government support and use of existing health infrastructure. This is to ensure an effective response to future health threats [26].

